Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC014782A_C01 KMC014782A_c01
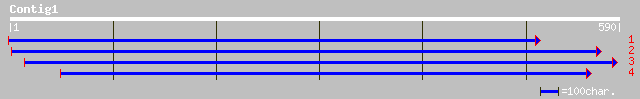
(590 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL93064.1|AC093180_11 putative ABC transporter [Oryza sativa... 79 6e-14
gb|AAM61469.1| putative ABC transporter [Arabidopsis thaliana] 77 2e-13
ref|NP_567001.1| ABC transporter family protein; protein id: At3... 77 2e-13
gb|AAL87178.1|AF480497_6 putative ABC transporter protein [Oryza... 67 2e-10
gb|AAL87694.1|AF479256_1 non-transporter ABC protein AbcF4 [Dict... 39 0.064
>gb|AAL93064.1|AC093180_11 putative ABC transporter [Oryza sativa (japonica cultivar-group)]
Length = 710
Score = 78.6 bits (192), Expect = 6e-14
Identities = 34/45 (75%), Positives = 41/45 (90%)
Frame = -1
Query: 590 ISRVCDDEERSQIWIVEDGTVRNFPGTFEDYKDDLLKEIKAEVDD 456
ISRVCDDE+RS+IW+VEDGTV F GTFEDYKD+LL+EIK EV++
Sbjct: 666 ISRVCDDEQRSEIWVVEDGTVNKFDGTFEDYKDELLEEIKKEVEE 710
>gb|AAM61469.1| putative ABC transporter [Arabidopsis thaliana]
Length = 723
Score = 77.0 bits (188), Expect = 2e-13
Identities = 34/45 (75%), Positives = 41/45 (90%)
Frame = -1
Query: 590 ISRVCDDEERSQIWIVEDGTVRNFPGTFEDYKDDLLKEIKAEVDD 456
ISRVC +EE+SQIW+VEDGTV FPGTFE+YK+DL +EIKAEVD+
Sbjct: 679 ISRVCAEEEKSQIWVVEDGTVNFFPGTFEEYKEDLQREIKAEVDE 723
>ref|NP_567001.1| ABC transporter family protein; protein id: At3g54540.1, supported
by cDNA: 120919., supported by cDNA: gi_15450889
[Arabidopsis thaliana] gi|11357152|pir||T47613 ABC
transporter-like protein - Arabidopsis thaliana
gi|7258357|emb|CAB77574.1| ABC transporter-like protein
[Arabidopsis thaliana] gi|15450890|gb|AAK96716.1| ABC
transporter-like protein [Arabidopsis thaliana]
Length = 723
Score = 77.0 bits (188), Expect = 2e-13
Identities = 34/45 (75%), Positives = 41/45 (90%)
Frame = -1
Query: 590 ISRVCDDEERSQIWIVEDGTVRNFPGTFEDYKDDLLKEIKAEVDD 456
ISRVC +EE+SQIW+VEDGTV FPGTFE+YK+DL +EIKAEVD+
Sbjct: 679 ISRVCAEEEKSQIWVVEDGTVNFFPGTFEEYKEDLQREIKAEVDE 723
>gb|AAL87178.1|AF480497_6 putative ABC transporter protein [Oryza sativa (japonica
cultivar-group)]
Length = 606
Score = 67.0 bits (162), Expect = 2e-10
Identities = 27/43 (62%), Positives = 37/43 (85%)
Frame = -1
Query: 590 ISRVCDDEERSQIWIVEDGTVRNFPGTFEDYKDDLLKEIKAEV 462
+SRVCDDEERS +W+V+DGTVR + GTF +Y+DDLL +I+ E+
Sbjct: 561 VSRVCDDEERSALWVVQDGTVRPYDGTFAEYRDDLLDDIRKEM 603
>gb|AAL87694.1|AF479256_1 non-transporter ABC protein AbcF4 [Dictyostelium discoideum]
Length = 1142
Score = 38.5 bits (88), Expect = 0.064
Identities = 16/29 (55%), Positives = 24/29 (82%), Gaps = 1/29 (3%)
Frame = -1
Query: 557 QIWIVE-DGTVRNFPGTFEDYKDDLLKEI 474
QIW+V+ DGT+ +PGTF DYK+++ +EI
Sbjct: 1107 QIWVVKKDGTIYLYPGTFMDYKNEISREI 1135
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 468,070,241
Number of Sequences: 1393205
Number of extensions: 9944689
Number of successful extensions: 22300
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 21618
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22287
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22569056698
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)