Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC014549A_C01 KMC014549A_c01
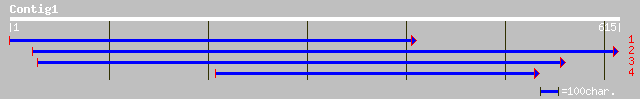
(613 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T05694 pathogenesis-related protein F20M13.220 - Arabidopsi... 130 1e-29
ref|NP_568046.1| putative thaumatin-like protein; protein id: At... 130 1e-29
gb|AAM64698.1| putative thaumatin-like protein [Arabidopsis thal... 130 1e-29
ref|NP_195325.1| thaumatin-like protein; protein id: At4g36010.1... 121 8e-27
ref|NP_195324.1| thaumatin-like protein; protein id: At4g36000.1... 115 5e-25
>pir||T05694 pathogenesis-related protein F20M13.220 - Arabidopsis thaliana
gi|4467153|emb|CAB37522.1| putative thaumatin-like
protein [Arabidopsis thaliana]
gi|7270849|emb|CAB80530.1| putative thaumatin-like
protein [Arabidopsis thaliana]
Length = 323
Score = 130 bits (327), Expect = 1e-29
Identities = 73/154 (47%), Positives = 95/154 (61%), Gaps = 25/154 (16%)
Frame = -1
Query: 613 PEYCCNGAFSNPSTCKPSVYSEMFKSACPKSYSYAYDDATSTFTCSGADYTITFCPSSSP 434
PEYCC+GA++ PS+C+PSVYSEMFK+ACP+SYSYAYDDATSTFTC+G DYT+TFCP SSP
Sbjct: 168 PEYCCSGAYATPSSCRPSVYSEMFKAACPRSYSYAYDDATSTFTCAGGDYTVTFCP-SSP 226
Query: 433 ILKSSTDSPPKAIDSDSGSSSSS-----------------------GSVGSGS--SSEQS 329
KS++ SPP DS S S S GS G+GS + ++
Sbjct: 227 SQKSTSYSPP-VTDSSSTSQGSDPVPGSDTGYAGQGQQTPGQGNVYGSQGTGSEMGTGET 285
Query: 328 AIASTSWMADMATATGGSSTRSRDLCWWMLLSFF 227
+ SWMA +A G ++R + +LL+ F
Sbjct: 286 MLQDGSWMAGLAM---GEASRPAGVSLTVLLAAF 316
>ref|NP_568046.1| putative thaumatin-like protein; protein id: At4g38660.1, supported
by cDNA: 32152., supported by cDNA: gi_17979354,
supported by cDNA: gi_20466020 [Arabidopsis thaliana]
gi|17979355|gb|AAL49903.1| putative thaumatin protein
[Arabidopsis thaliana] gi|20466021|gb|AAM20232.1|
putative thaumatin [Arabidopsis thaliana]
Length = 345
Score = 130 bits (327), Expect = 1e-29
Identities = 73/154 (47%), Positives = 95/154 (61%), Gaps = 25/154 (16%)
Frame = -1
Query: 613 PEYCCNGAFSNPSTCKPSVYSEMFKSACPKSYSYAYDDATSTFTCSGADYTITFCPSSSP 434
PEYCC+GA++ PS+C+PSVYSEMFK+ACP+SYSYAYDDATSTFTC+G DYT+TFCP SSP
Sbjct: 190 PEYCCSGAYATPSSCRPSVYSEMFKAACPRSYSYAYDDATSTFTCAGGDYTVTFCP-SSP 248
Query: 433 ILKSSTDSPPKAIDSDSGSSSSS-----------------------GSVGSGS--SSEQS 329
KS++ SPP DS S S S GS G+GS + ++
Sbjct: 249 SQKSTSYSPP-VTDSSSTSQGSDPVPGSDTGYAGQGQQTPGQGNVYGSQGTGSEMGTGET 307
Query: 328 AIASTSWMADMATATGGSSTRSRDLCWWMLLSFF 227
+ SWMA +A G ++R + +LL+ F
Sbjct: 308 MLQDGSWMAGLAM---GEASRPAGVSLTVLLAAF 338
>gb|AAM64698.1| putative thaumatin-like protein [Arabidopsis thaliana]
Length = 345
Score = 130 bits (327), Expect = 1e-29
Identities = 73/154 (47%), Positives = 95/154 (61%), Gaps = 25/154 (16%)
Frame = -1
Query: 613 PEYCCNGAFSNPSTCKPSVYSEMFKSACPKSYSYAYDDATSTFTCSGADYTITFCPSSSP 434
PEYCC+GA++ PS+C+PSVYSEMFK+ACP+SYSYAYDDATSTFTC+G DYT+TFCP SSP
Sbjct: 190 PEYCCSGAYATPSSCRPSVYSEMFKAACPRSYSYAYDDATSTFTCAGGDYTVTFCP-SSP 248
Query: 433 ILKSSTDSPPKAIDSDSGSSSSS-----------------------GSVGSGS--SSEQS 329
KS++ SPP DS S S S GS G+GS + ++
Sbjct: 249 SQKSTSYSPP-VTDSSSTSQGSDPVPGSDTGYAGQGQQTPGQGNVYGSQGTGSEMGTGET 307
Query: 328 AIASTSWMADMATATGGSSTRSRDLCWWMLLSFF 227
+ SWMA +A G ++R + +LL+ F
Sbjct: 308 MLQDGSWMAGLAM---GEASRPAGVSLTVLLAAF 338
>ref|NP_195325.1| thaumatin-like protein; protein id: At4g36010.1, supported by cDNA:
gi_13430505, supported by cDNA: gi_21280948 [Arabidopsis
thaliana] gi|7442167|pir||T05493 pathogenesis-related
protein 19K4.140 - Arabidopsis thaliana
gi|3036805|emb|CAA18495.1| thaumatin-like protein
[Arabidopsis thaliana] gi|7270553|emb|CAB81510.1|
thaumatin-like protein [Arabidopsis thaliana]
gi|13430506|gb|AAK25875.1|AF360165_1 putative thaumatin
protein [Arabidopsis thaliana]
gi|21280949|gb|AAM44961.1| putative thaumatin protein
[Arabidopsis thaliana]
Length = 301
Score = 121 bits (303), Expect = 8e-27
Identities = 56/100 (56%), Positives = 71/100 (71%), Gaps = 1/100 (1%)
Frame = -1
Query: 613 PEYCCNGAFSNPSTCKPSVYSEMFKSACPKSYSYAYDDATSTFTCSGADYTITFCPSSSP 434
PEYCC+GAF P TCKPS YS+ FK+ACP++YSYAYDD TSTFTC GADY ITFCPS +P
Sbjct: 195 PEYCCSGAFGTPDTCKPSEYSQFFKNACPRAYSYAYDDGTSTFTCGGADYVITFCPSPNP 254
Query: 433 ILKSSTDS-PPKAIDSDSGSSSSSGSVGSGSSSEQSAIAS 317
+KS+T P A+ S ++S ++ + S A+AS
Sbjct: 255 SVKSATKGVQPVAVSYSKASPNASPTLSAVFSIGVLAVAS 294
>ref|NP_195324.1| thaumatin-like protein; protein id: At4g36000.1 [Arabidopsis
thaliana] gi|7488366|pir||T05492 thaumatin homolog
T19K4.130 - Arabidopsis thaliana
gi|3036804|emb|CAA18494.1| thaumatin-like protein
[Arabidopsis thaliana] gi|7270552|emb|CAB81509.1|
thaumatin-like protein [Arabidopsis thaliana]
Length = 190
Score = 115 bits (288), Expect = 5e-25
Identities = 48/59 (81%), Positives = 55/59 (92%)
Frame = -1
Query: 613 PEYCCNGAFSNPSTCKPSVYSEMFKSACPKSYSYAYDDATSTFTCSGADYTITFCPSSS 437
PEYCC+GA+ +P +CKPS+YS++FKSACPKSYSYAYDDATSTFTCSGADYTITFCP S
Sbjct: 131 PEYCCSGAYGSPDSCKPSMYSQVFKSACPKSYSYAYDDATSTFTCSGADYTITFCPKLS 189
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 550,265,767
Number of Sequences: 1393205
Number of extensions: 12418563
Number of successful extensions: 99414
Number of sequences better than 10.0: 874
Number of HSP's better than 10.0 without gapping: 58133
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 82799
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24568846532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)