Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
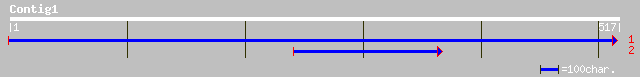
Query= KMC014323A_C01 KMC014323A_c01
(517 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|Q9FV53|DEFM_ARATH Peptide deformylase, mitochondrial precurso... 80 2e-14
ref|NP_563974.1| expressed protein; protein id: At1g15390.1, sup... 80 2e-14
sp|Q9FUZ0|DEFM_LYCES Peptide deformylase, mitochondrial precurso... 70 1e-11
ref|NP_406383.1| histidyl-tRNA synthetase [Yersinia pestis] gi|2... 33 1.9
ref|NP_668676.1| histidine tRNA synthetase [Yersinia pestis KIM]... 32 3.3
>sp|Q9FV53|DEFM_ARATH Peptide deformylase, mitochondrial precursor (PDF) (Polypeptide
deformylase) gi|25518549|pir||E86288 hypothetical
protein F9L1.34 - Arabidopsis thaliana
gi|5103837|gb|AAD39667.1|AC007591_32 Simalar to
gi|4377403 Polypeptide Deformylase from Chlamydia
pneumoniae genome gb|AE001687. [Arabidopsis thaliana]
gi|13605760|gb|AAK32873.1|AF361861_1 At1g15390/F9L1_34
[Arabidopsis thaliana] gi|22136570|gb|AAM91071.1|
At1g15390/F9L1_34 [Arabidopsis thaliana]
Length = 259
Score = 79.7 bits (195), Expect = 2e-14
Identities = 34/39 (87%), Positives = 37/39 (94%)
Frame = -1
Query: 517 ECDHLDGTLYVDKMVPRTFRTVDNLNLPLGQGCPKLGPR 401
ECDHLDG LYVDKMVPRTFRTVDNL+LPL +GCPKLGP+
Sbjct: 221 ECDHLDGNLYVDKMVPRTFRTVDNLDLPLAEGCPKLGPQ 259
>ref|NP_563974.1| expressed protein; protein id: At1g15390.1, supported by cDNA:
gi_11320951 [Arabidopsis thaliana]
gi|11320952|gb|AAG33973.1|AF250959_1 peptide
deformylase-like protein [Arabidopsis thaliana]
Length = 269
Score = 79.7 bits (195), Expect = 2e-14
Identities = 34/39 (87%), Positives = 37/39 (94%)
Frame = -1
Query: 517 ECDHLDGTLYVDKMVPRTFRTVDNLNLPLGQGCPKLGPR 401
ECDHLDG LYVDKMVPRTFRTVDNL+LPL +GCPKLGP+
Sbjct: 231 ECDHLDGNLYVDKMVPRTFRTVDNLDLPLAEGCPKLGPQ 269
>sp|Q9FUZ0|DEFM_LYCES Peptide deformylase, mitochondrial precursor (PDF) (Polypeptide
deformylase) gi|11320968|gb|AAG33981.1|AF271258_1
peptide deformylase-like protein [Lycopersicon
esculentum]
Length = 277
Score = 70.1 bits (170), Expect = 1e-11
Identities = 31/37 (83%), Positives = 33/37 (88%)
Frame = -1
Query: 517 ECDHLDGTLYVDKMVPRTFRTVDNLNLPLGQGCPKLG 407
E DHLDGTLYVDKM PRTFRTV+NL+LPL GCPKLG
Sbjct: 239 EYDHLDGTLYVDKMAPRTFRTVENLDLPLAAGCPKLG 275
>ref|NP_406383.1| histidyl-tRNA synthetase [Yersinia pestis] gi|25293367|pir||AF0350
histidine-tRNA ligase (EC 6.1.1.21) [imported] -
Yersinia pestis (strain CO92)
gi|15980845|emb|CAC92129.1| histidyl-tRNA synthetase
[Yersinia pestis CO92]
Length = 424
Score = 33.1 bits (74), Expect = 1.9
Identities = 12/31 (38%), Positives = 20/31 (63%)
Frame = -2
Query: 387 GFCGCLR*GLQHRFLVQVSVKLTFVGPIFRF 295
G GC+R G++H L +L ++GP+FR+
Sbjct: 84 GTAGCVRAGIEHGLLYNQEQRLWYIGPMFRY 114
>ref|NP_668676.1| histidine tRNA synthetase [Yersinia pestis KIM]
gi|21958124|gb|AAM84927.1|AE013738_4 histidine tRNA
synthetase [Yersinia pestis KIM]
Length = 424
Score = 32.3 bits (72), Expect = 3.3
Identities = 12/31 (38%), Positives = 19/31 (60%)
Frame = -2
Query: 387 GFCGCLR*GLQHRFLVQVSVKLTFVGPIFRF 295
G GC R G++H L +L ++GP+FR+
Sbjct: 84 GTAGCARAGIEHGLLYNQEQRLWYIGPMFRY 114
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 432,520,211
Number of Sequences: 1393205
Number of extensions: 8735012
Number of successful extensions: 24093
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 23258
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24090
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16154357632
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)