Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC014182A_C01 KMC014182A_c01
(800 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
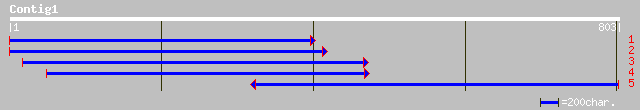
ref|NP_177692.1| unknown protein; protein id: At1g75620.1 [Arabi... 197 1e-49
pir||D86332 hypothetical protein F6F9.4 [imported] - Arabidopsis... 193 2e-48
ref|NP_173419.1| unknown protein; protein id: At1g19900.1, suppo... 193 2e-48
pir||H86278 F14L17.20 protein - Arabidopsis thaliana gi|7262685|... 166 3e-40
ref|NP_172895.1| hypothetical protein; protein id: At1g14430.1 [... 163 3e-39
>ref|NP_177692.1| unknown protein; protein id: At1g75620.1 [Arabidopsis thaliana]
gi|9369366|gb|AAF87115.1|AC006434_11 F10A5.18
[Arabidopsis thaliana]
Length = 547
Score = 197 bits (502), Expect = 1e-49
Identities = 106/181 (58%), Positives = 134/181 (73%), Gaps = 1/181 (0%)
Frame = -3
Query: 780 PGRSQFELQNPTKTPWMYHSTAVLLRDGRVLVAGSNPHEYYNFSSVLFPTELSLEAFSPP 601
P S+FE+QNP+ P MYHS A LLRDGR+LV GSNPH +YNF+ VLFPTEL LEAFSP
Sbjct: 372 PVGSRFEVQNPSTIPRMYHSIATLLRDGRILVGGSNPHAFYNFTGVLFPTELRLEAFSPS 431
Query: 600 YLEPQYTNVRPRIVSPTPQAQTKLKYGQGLRIRFQVTALLAAPDSVSMTMLAPPFNTHSF 421
YL+ +Y+++RP IV P P QT + YG+ LR+RF V+ + +P V +TML P F THSF
Sbjct: 432 YLDTKYSSLRPSIVDPRP--QTTVNYGRVLRLRFIVSGRVKSP--VKVTMLFPSFTTHSF 487
Query: 420 SMNQRLLVLEPNGLKDVGES-IHEVEVTAPGSATLAPPGFYLVFVVHQRRRIPSEGIWVQ 244
SM+QRLLVL+ +G S I+EV V P SA LAPPG+Y+VFVV+Q IPSEG+WV+
Sbjct: 488 SMHQRLLVLDHVISFKLGISKIYEVRVRTPSSAILAPPGYYMVFVVNQ--DIPSEGLWVR 545
Query: 243 I 241
+
Sbjct: 546 L 546
>pir||D86332 hypothetical protein F6F9.4 [imported] - Arabidopsis thaliana
gi|10086483|gb|AAG12543.1|AC007797_3 Unknown Protein
[Arabidopsis thaliana]
Length = 504
Score = 193 bits (491), Expect = 2e-48
Identities = 99/181 (54%), Positives = 133/181 (72%), Gaps = 1/181 (0%)
Frame = -3
Query: 780 PGRSQFELQNPTKTPWMYHSTAVLLRDGRVLVAGSNPHEYYNFSSVLFPTELSLEAFSPP 601
P S+FE PT P MYHS A+LLRDGRVLV GSNPH +YN++ VLFPTELSLEAFSP
Sbjct: 329 PVGSRFESLRPTTIPRMYHSAAILLRDGRVLVGGSNPHAFYNYTGVLFPTELSLEAFSPV 388
Query: 600 YLEPQYTNVRPRIVSPTPQAQTKLKYGQGLRIRFQVTALLAAPDSVSMTMLAPPFNTHSF 421
YL+ +++N+RP+I+SP PQ+ +KYG L+++F VT + P V TM+ P F THSF
Sbjct: 389 YLQREFSNLRPKIISPEPQSM--IKYGTNLKLKFSVTGEVTTPAKV--TMVFPTFTTHSF 444
Query: 420 SMNQRLLVLEPNGLKDVGES-IHEVEVTAPGSATLAPPGFYLVFVVHQRRRIPSEGIWVQ 244
+MNQR+LVL+ G+S ++EV+V P SA +A PG+Y++FVV+Q IPSEG+WV+
Sbjct: 445 AMNQRVLVLDNVKFTRKGKSPMYEVQVRTPRSANIAWPGYYMIFVVNQ--DIPSEGVWVK 502
Query: 243 I 241
+
Sbjct: 503 L 503
>ref|NP_173419.1| unknown protein; protein id: At1g19900.1, supported by cDNA:
gi_16604506 [Arabidopsis thaliana]
gi|16604507|gb|AAL24259.1| At1g19900/F6F9_4 [Arabidopsis
thaliana]
Length = 548
Score = 193 bits (491), Expect = 2e-48
Identities = 99/181 (54%), Positives = 133/181 (72%), Gaps = 1/181 (0%)
Frame = -3
Query: 780 PGRSQFELQNPTKTPWMYHSTAVLLRDGRVLVAGSNPHEYYNFSSVLFPTELSLEAFSPP 601
P S+FE PT P MYHS A+LLRDGRVLV GSNPH +YN++ VLFPTELSLEAFSP
Sbjct: 373 PVGSRFESLRPTTIPRMYHSAAILLRDGRVLVGGSNPHAFYNYTGVLFPTELSLEAFSPV 432
Query: 600 YLEPQYTNVRPRIVSPTPQAQTKLKYGQGLRIRFQVTALLAAPDSVSMTMLAPPFNTHSF 421
YL+ +++N+RP+I+SP PQ+ +KYG L+++F VT + P V TM+ P F THSF
Sbjct: 433 YLQREFSNLRPKIISPEPQSM--IKYGTNLKLKFSVTGEVTTPAKV--TMVFPTFTTHSF 488
Query: 420 SMNQRLLVLEPNGLKDVGES-IHEVEVTAPGSATLAPPGFYLVFVVHQRRRIPSEGIWVQ 244
+MNQR+LVL+ G+S ++EV+V P SA +A PG+Y++FVV+Q IPSEG+WV+
Sbjct: 489 AMNQRVLVLDNVKFTRKGKSPMYEVQVRTPRSANIAWPGYYMIFVVNQ--DIPSEGVWVK 546
Query: 243 I 241
+
Sbjct: 547 L 547
>pir||H86278 F14L17.20 protein - Arabidopsis thaliana
gi|7262685|gb|AAF43943.1|AC012188_20 Weak similarity to
glyoxal oxidase (glx2) from Phanerochaete chrysosporium
gb|L47287. [Arabidopsis thaliana]
Length = 564
Score = 166 bits (421), Expect = 3e-40
Identities = 85/186 (45%), Positives = 119/186 (63%)
Frame = -3
Query: 798 YLQNKYPGRSQFELQNPTKTPWMYHSTAVLLRDGRVLVAGSNPHEYYNFSSVLFPTELSL 619
YL + +FE+ PT+ P MYHS ++LL DGRVLV GSNPH YNF++ +PTELSL
Sbjct: 383 YLPEEPDQTRRFEILTPTRIPRMYHSASLLLSDGRVLVGGSNPHRNYNFTARPYPTELSL 442
Query: 618 EAFSPPYLEPQYTNVRPRIVSPTPQAQTKLKYGQGLRIRFQVTALLAAPDSVSMTMLAPP 439
EA+ P YL+PQY VRP I+ T + + YGQ + F + A VS+ ++AP
Sbjct: 443 EAYLPRYLDPQYARVRPTII--TVELAGNMLYGQAFAVTFAIPAFGMFDGGVSVRLVAPS 500
Query: 438 FNTHSFSMNQRLLVLEPNGLKDVGESIHEVEVTAPGSATLAPPGFYLVFVVHQRRRIPSE 259
F+THS +MNQRLLVL + + ++ +V P ++ +APPG+Y++FVVH R IPS
Sbjct: 501 FSTHSTAMNQRLLVLRVRRVSQLSVFAYKADVDGPTNSYVAPPGYYMMFVVH--RGIPSV 558
Query: 258 GIWVQI 241
+WV+I
Sbjct: 559 AVWVKI 564
>ref|NP_172895.1| hypothetical protein; protein id: At1g14430.1 [Arabidopsis
thaliana]
Length = 849
Score = 163 bits (412), Expect = 3e-39
Identities = 83/183 (45%), Positives = 116/183 (63%)
Frame = -3
Query: 798 YLQNKYPGRSQFELQNPTKTPWMYHSTAVLLRDGRVLVAGSNPHEYYNFSSVLFPTELSL 619
YL + +FE+ PT+ P MYHS ++LL DGRVLV GSNPH YNF++ +PTELSL
Sbjct: 383 YLPEEPDQTRRFEILTPTRIPRMYHSASLLLSDGRVLVGGSNPHRNYNFTARPYPTELSL 442
Query: 618 EAFSPPYLEPQYTNVRPRIVSPTPQAQTKLKYGQGLRIRFQVTALLAAPDSVSMTMLAPP 439
EA+ P YL+PQY VRP I+ T + + YGQ + F + A VS+ ++AP
Sbjct: 443 EAYLPRYLDPQYARVRPTII--TVELAGNMLYGQAFAVTFAIPAFGMFDGGVSVRLVAPS 500
Query: 438 FNTHSFSMNQRLLVLEPNGLKDVGESIHEVEVTAPGSATLAPPGFYLVFVVHQRRRIPSE 259
F+THS +MNQRLLVL + + ++ +V P ++ +APPG+Y++FVVH R IPS
Sbjct: 501 FSTHSTAMNQRLLVLRVRRVSQLSVFAYKADVDGPTNSYVAPPGYYMMFVVH--RGIPSV 558
Query: 258 GIW 250
+W
Sbjct: 559 AVW 561
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 763,339,826
Number of Sequences: 1393205
Number of extensions: 18776892
Number of successful extensions: 57145
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 54245
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 57085
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40616159090
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)