Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC013962A_C01 KMC013962A_c01
(559 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
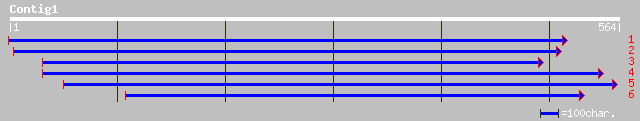
gb|AAG34797.1|AF243362_1 glutathione S-transferase GST 7 [Glycin... 149 2e-35
gb|AAG34798.1|AF243363_1 glutathione S-transferase GST 8 [Glycin... 147 1e-34
gb|AAO69664.1| glutathione S-transferase [Phaseolus acutifolius] 147 1e-34
pir||T10825 auxin-induced protein (clone MII-4) - mung bean (fra... 146 2e-34
sp|P32110|GTX6_SOYBN PROBABLE GLUTATHIONE S-TRANSFERASE (HEAT SH... 142 4e-33
>gb|AAG34797.1|AF243362_1 glutathione S-transferase GST 7 [Glycine max]
Length = 225
Score = 149 bits (376), Expect = 2e-35
Identities = 75/116 (64%), Positives = 91/116 (77%), Gaps = 5/116 (4%)
Frame = -2
Query: 558 AAW----TANQEEREKGIQESVEALQFLEKELKHK-FFGGETIGLVDIAGAFLAFWLPVI 394
AAW T +++EREK ++ES EALQFLE ELK K FFGGE IGLVDIA F+AFW+P+I
Sbjct: 111 AAWKYIYTVDEKEREKNVEESYEALQFLENELKDKKFFGGEEIGLVDIAAVFIAFWIPII 170
Query: 393 EESTGFSLLTSEKFPKLYKWSQDFINHPVVKENLPPRDKLLGFFKGRYAASINASK 226
+E G L TSEKFPKLYKWSQ+FINHPVVK+ LPPRD+L F+K + S++ASK
Sbjct: 171 QEVLGLKLFTSEKFPKLYKWSQEFINHPVVKQVLPPRDQLFAFYKACH-ESLSASK 225
>gb|AAG34798.1|AF243363_1 glutathione S-transferase GST 8 [Glycine max]
Length = 225
Score = 147 bits (370), Expect = 1e-34
Identities = 67/108 (62%), Positives = 87/108 (80%), Gaps = 5/108 (4%)
Frame = -2
Query: 558 AAW----TANQEEREKGIQESVEALQFLEKELKHK-FFGGETIGLVDIAGAFLAFWLPVI 394
AAW TA+++EREK ++E++EALQFLE E+K K FFGGE IGLVDIA ++AFW+P++
Sbjct: 111 AAWKSVFTADEKEREKNVEEAIEALQFLENEIKDKKFFGGEEIGLVDIAAVYIAFWVPMV 170
Query: 393 EESTGFSLLTSEKFPKLYKWSQDFINHPVVKENLPPRDKLLGFFKGRY 250
+E G L TSEKFPKL+ WSQ+F+NHP+VKE+LPPRD + FFKG Y
Sbjct: 171 QEIAGLELFTSEKFPKLHNWSQEFLNHPIVKESLPPRDPVFSFFKGLY 218
>gb|AAO69664.1| glutathione S-transferase [Phaseolus acutifolius]
Length = 225
Score = 147 bits (370), Expect = 1e-34
Identities = 68/112 (60%), Positives = 91/112 (80%), Gaps = 1/112 (0%)
Frame = -2
Query: 558 AAWTANQEEREKGIQESVEALQFLEKELKHK-FFGGETIGLVDIAGAFLAFWLPVIEEST 382
+ +TA+++EREK + E+ E+LQFLE E+ K FFGGE +GLVDIA ++AFW+P+++E
Sbjct: 115 SVFTADEKEREKNVAEASESLQFLENEIADKKFFGGEELGLVDIAAVYVAFWIPLVQEIA 174
Query: 381 GFSLLTSEKFPKLYKWSQDFINHPVVKENLPPRDKLLGFFKGRYAASINASK 226
G LLTSEKFP LYKWSQ+F++HP+VKE+LPPRD + GFFKGRY S+ ASK
Sbjct: 175 GLELLTSEKFPNLYKWSQEFVSHPIVKESLPPRDPVFGFFKGRY-ESLFASK 225
>pir||T10825 auxin-induced protein (clone MII-4) - mung bean (fragment)
gi|1184123|gb|AAA87183.1| auxin-induced protein [Vigna
radiata]
Length = 230
Score = 146 bits (368), Expect = 2e-34
Identities = 65/104 (62%), Positives = 85/104 (81%), Gaps = 1/104 (0%)
Frame = -2
Query: 558 AAWTANQEEREKGIQESVEALQFLEKELKHK-FFGGETIGLVDIAGAFLAFWLPVIEEST 382
+ +T +++EREK I E+ E+LQFLE E+K K FFGGE +GLVDIA ++AFW+P+I+E
Sbjct: 120 SVFTVDEKEREKNIAETYESLQFLENEIKEKKFFGGEELGLVDIAAVYVAFWIPLIQEIA 179
Query: 381 GFSLLTSEKFPKLYKWSQDFINHPVVKENLPPRDKLLGFFKGRY 250
G LLTSEKFP LY+WSQ+F+NHP+VKE+LPPRD + FFKGRY
Sbjct: 180 GLELLTSEKFPNLYRWSQEFLNHPIVKESLPPRDPVFAFFKGRY 223
>sp|P32110|GTX6_SOYBN PROBABLE GLUTATHIONE S-TRANSFERASE (HEAT SHOCK PROTEIN 26A) (G2-4)
gi|99912|pir||A33654 heat shock protein 26A - soybean
gi|169981|gb|AAA33973.1| Gmhsp26-A
Length = 225
Score = 142 bits (357), Expect = 4e-33
Identities = 68/112 (60%), Positives = 85/112 (75%), Gaps = 1/112 (0%)
Frame = -2
Query: 558 AAWTANQEEREKGIQESVEALQFLEKELKHK-FFGGETIGLVDIAGAFLAFWLPVIEEST 382
+ +T +++EREK ++E+ EALQFLE ELK K FFGGE GLVDIA F+AFW+P+ +E
Sbjct: 115 SVFTVDEKEREKNVEETYEALQFLENELKDKKFFGGEEFGLVDIAAVFIAFWIPIFQEIA 174
Query: 381 GFSLLTSEKFPKLYKWSQDFINHPVVKENLPPRDKLLGFFKGRYAASINASK 226
G L TSEKFP LYKWSQ+F+NHP V E LPPRD L +FK RY S++ASK
Sbjct: 175 GLQLFTSEKFPILYKWSQEFLNHPFVHEVLPPRDPLFAYFKARY-ESLSASK 225
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 471,943,288
Number of Sequences: 1393205
Number of extensions: 9427225
Number of successful extensions: 23998
Number of sequences better than 10.0: 214
Number of HSP's better than 10.0 without gapping: 23411
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23931
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)