Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
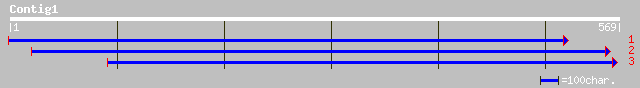
Query= KMC013629A_C01 KMC013629A_c01
(569 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_188028.1| hypothetical protein; protein id: At3g14120.1 [... 167 1e-40
gb|AAM98082.1| AT3g14120/MAG2_7 [Arabidopsis thaliana] 167 1e-40
dbj|BAB02976.1| gene_id:MAG2.8~similar to unknown protein~sp|P52... 146 2e-34
ref|ZP_00059709.1| hypothetical protein [Clostridium thermocellu... 36 0.30
gb|EAA19555.1| hypothetical protein [Plasmodium yoelii yoelii] 35 0.51
>ref|NP_188028.1| hypothetical protein; protein id: At3g14120.1 [Arabidopsis thaliana]
Length = 1079
Score = 167 bits (422), Expect = 1e-40
Identities = 81/111 (72%), Positives = 95/111 (84%)
Frame = -2
Query: 562 ELPRFQAGITMEISRLDAWYSDKSGTLECPATYIVKGLCRRCCLPEVILRSMQVSVSLMG 383
EL RFQAG+TM+ISRLDAWYS K G+LE PATYIV+GLCRRCCLPE++LRSMQVSVSLM
Sbjct: 965 ELTRFQAGVTMDISRLDAWYSSKEGSLETPATYIVRGLCRRCCLPELVLRSMQVSVSLME 1024
Query: 382 SGVLPDCHDTLVELVGSPETQFVHLFSQQQLQEFLLFEREYSICKMELLEE 230
SG P+ HD L+ELV S ET F+ LFS+QQLQEF+LFEREY + ++EL EE
Sbjct: 1025 SGNPPEDHDELIELVASDETGFLSLFSRQQLQEFMLFEREYRMSQLELQEE 1075
>gb|AAM98082.1| AT3g14120/MAG2_7 [Arabidopsis thaliana]
Length = 1101
Score = 167 bits (422), Expect = 1e-40
Identities = 81/111 (72%), Positives = 95/111 (84%)
Frame = -2
Query: 562 ELPRFQAGITMEISRLDAWYSDKSGTLECPATYIVKGLCRRCCLPEVILRSMQVSVSLMG 383
EL RFQAG+TM+ISRLDAWYS K G+LE PATYIV+GLCRRCCLPE++LRSMQVSVSLM
Sbjct: 987 ELTRFQAGVTMDISRLDAWYSSKEGSLETPATYIVRGLCRRCCLPELVLRSMQVSVSLME 1046
Query: 382 SGVLPDCHDTLVELVGSPETQFVHLFSQQQLQEFLLFEREYSICKMELLEE 230
SG P+ HD L+ELV S ET F+ LFS+QQLQEF+LFEREY + ++EL EE
Sbjct: 1047 SGNPPEDHDELIELVASDETGFLSLFSRQQLQEFMLFEREYRMSQLELQEE 1097
>dbj|BAB02976.1| gene_id:MAG2.8~similar to unknown protein~sp|P52590 [Arabidopsis
thaliana]
Length = 1158
Score = 146 bits (369), Expect = 2e-34
Identities = 71/94 (75%), Positives = 81/94 (85%)
Frame = -2
Query: 568 KGELPRFQAGITMEISRLDAWYSDKSGTLECPATYIVKGLCRRCCLPEVILRSMQVSVSL 389
KGEL RFQAG+TM+ISRLDAWYS K G+LE PATYIV+GLCRRCCLPE++LRSMQVSVSL
Sbjct: 996 KGELTRFQAGVTMDISRLDAWYSSKEGSLETPATYIVRGLCRRCCLPELVLRSMQVSVSL 1055
Query: 388 MGSGVLPDCHDTLVELVGSPETQFVHLFSQQQLQ 287
M SG P+ HD L+ELV S ET F+ LFS+QQLQ
Sbjct: 1056 MESGNPPEDHDELIELVASDETGFLSLFSRQQLQ 1089
>ref|ZP_00059709.1| hypothetical protein [Clostridium thermocellum ATCC 27405]
Length = 370
Score = 36.2 bits (82), Expect = 0.30
Identities = 32/155 (20%), Positives = 76/155 (48%), Gaps = 11/155 (7%)
Frame = -2
Query: 442 RCCLPEVILRSMQVSVSLMGSGVLPDCHDTLVELVGSPETQFVHLFSQQQLQEFLLFERE 263
R +P V+L+++ + +GS + P +G+P+ +++ SQ + EF+LF
Sbjct: 119 RLVIPVVVLQTIAAN---LGSMLTP---------IGNPQNLYLYNLSQMGILEFVLFMLP 166
Query: 262 YSICKMELL-----------EE*ILSFEQNLIVSFFQSHRFVLVF*TFFGVS*ATVLKLY 116
Y++ LL E ++ E N+ +S ++ ++++F +S A +L
Sbjct: 167 YTMVSGLLLVISLLLIKGKHEAVVIKEETNMRLSLKKNIIYLILF-ALSLLSVAKILHYM 225
Query: 115 IVICLVIILL*VEKREYEKLCA*ACLVNYYFYLVF 11
+V+ LV++++ + +++ K L+ + + +F
Sbjct: 226 VVLILVLLVVFIMEKDVLKSVDYCLLLTFICFFIF 260
>gb|EAA19555.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 1011
Score = 35.4 bits (80), Expect = 0.51
Identities = 19/73 (26%), Positives = 43/73 (58%)
Frame = +3
Query: 69 SLFSTQSKIITRQITIYNFKTVAQDTPKNVQNTKTNLWLWKKLTIKFCSKDKIHSSKSSI 248
S+F +++ + +++I++ K D+ ++ N+K +++ +K KF K+ I SK+SI
Sbjct: 549 SIFDSKNSLFGSKMSIFDSKMSIFDSKMSIFNSKNSIFKFKNNIFKF--KNSIFDSKNSI 606
Query: 249 LQIEYSLSNNKNS 287
+ S+ ++K S
Sbjct: 607 FDSKNSIFDSKKS 619
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,459,469
Number of Sequences: 1393205
Number of extensions: 10620384
Number of successful extensions: 30100
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 28907
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30082
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20956655091
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)