Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC013520A_C01 KMC013520A_c01
(1083 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
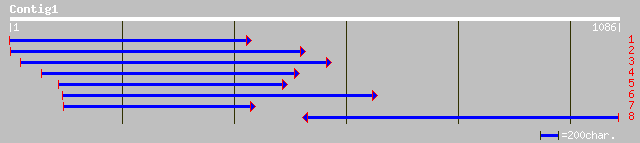
gb|AAM52321.1|AC105363_10 Putative nascent polypeptide associate... 250 1e-70
ref|NP_190516.1| alpha NAC-like protein; protein id: At3g49470.1... 231 1e-65
gb|AAK48972.1|AF370545_1 alpha NAC-like protein [Arabidopsis tha... 230 3e-65
ref|NP_187845.1| nascent polypeptide associated complex alpha ch... 208 1e-56
dbj|BAB89723.1| putative nascent polypeptide associated complex ... 207 1e-56
>gb|AAM52321.1|AC105363_10 Putative nascent polypeptide associated complex alpha chain [Oryza
sativa (japonica cultivar-group)]
gi|29367555|gb|AAO72639.1| putative nascent
polypeptide-associated complex alpha chain [Oryza sativa
(japonica cultivar-group)]
Length = 221
Score = 250 bits (638), Expect(2) = 1e-70
Identities = 138/210 (65%), Positives = 158/210 (74%)
Frame = -2
Query: 998 VSQNMAPGPVVDASSEAEALEQVLSAEETTLKKKPQPQEDDAPMVEDVKDDDKDEADDDE 819
VS+ AP +A E+ A V +E K +P EDDAP+VED KDDD DE DDD
Sbjct: 2 VSEQTAPVATAEAELESSAAPPV-KPDEAAAKAQP---EDDAPVVEDAKDDDDDEEDDD- 56
Query: 818 DEDDDDEDDDKEDGALGGSEGSKQSRSEKKSRKARLKLGLKPVTGVSRVTIKRTKNILFS 639
DD+DED+D E GA+G +EGSKQSRSEKKSRKA +KLG+KPVTGVSR+TIKR KNILF
Sbjct: 57 --DDEDEDEDGEQGAIG-NEGSKQSRSEKKSRKAMMKLGMKPVTGVSRITIKRAKNILFV 113
Query: 638 ISKPDVFKSPNSETYVICGEAKIEDLSSQLQTQAAQQFRMPDMGSVMAKQDQGTAADGGQ 459
+SKPDVFKSP SETYVI GEAKIEDLSSQLQ QAAQQFRM D+ VM+K D AA
Sbjct: 114 VSKPDVFKSPTSETYVIFGEAKIEDLSSQLQAQAAQQFRMQDLSKVMSKPDAAAAAPA-- 171
Query: 458 PEEEEEVDETGVEPHDIDLVMTQAGVSQGQ 369
+EEEEVDETG+EP DIDLVMTQA VS+ +
Sbjct: 172 -DEEEEVDETGIEPRDIDLVMTQASVSRAK 200
Score = 40.0 bits (92), Expect(2) = 1e-70
Identities = 19/23 (82%), Positives = 20/23 (86%)
Frame = -3
Query: 376 RGKAVKALKTHNGDIVGAIMELT 308
R KAVKALK H+GDIV AIMELT
Sbjct: 198 RAKAVKALKAHDGDIVSAIMELT 220
>ref|NP_190516.1| alpha NAC-like protein; protein id: At3g49470.1, supported by cDNA:
gi_13899100, supported by cDNA: gi_18377569 [Arabidopsis
thaliana] gi|11281614|pir||T46225 alpha NAC-like protein
- Arabidopsis thaliana gi|6561948|emb|CAB62452.1| alpha
NAC-like protein [Arabidopsis thaliana]
gi|12324452|gb|AAG52192.1|AC012329_19 putative alpha
NAC; 61864-63065 [Arabidopsis thaliana]
Length = 217
Score = 231 bits (589), Expect(2) = 1e-65
Identities = 132/208 (63%), Positives = 156/208 (74%), Gaps = 2/208 (0%)
Frame = -2
Query: 986 MAPGP-VVDASSEAEALEQVLSAEETTLKKKPQPQEDDAPMVEDVKDDDKDEADDDEDED 810
M+P P VV S++ + + ++A L+KK Q E P+VEDVKDD+ D+ DD+E+ED
Sbjct: 1 MSPPPAVVTESADGQPEQPPVTAIAEELEKKLQTDE---PIVEDVKDDEDDDDDDEEEED 57
Query: 809 DDDEDDDKEDGALGGSEGSKQSRSEKKSRKARLKLGLKPVTGVSRVTIKRTKNILFSISK 630
DD A G S SKQSRSEKKSRKA LKLG+KPVTGVSRVTIKRTKN+LF ISK
Sbjct: 58 DD---------AQGVSGSSKQSRSEKKSRKAMLKLGMKPVTGVSRVTIKRTKNVLFFISK 108
Query: 629 PDVFKSPNSETYVICGEAKIEDLSSQLQTQAAQQFRMPDMGSVMAKQDQGTAADGGQPEE 450
PDVFKSP+SETYVI GEAKIEDLSSQLQTQAAQQFRMP++G+ + + TA Q EE
Sbjct: 109 PDVFKSPHSETYVIFGEAKIEDLSSQLQTQAAQQFRMPEIGATSQRAEASTATVEAQVEE 168
Query: 449 -EEEVDETGVEPHDIDLVMTQAGVSQGQ 369
EEE+DETGVE DIDLVMTQAGVS+ +
Sbjct: 169 DEEEIDETGVEARDIDLVMTQAGVSRSK 196
Score = 42.4 bits (98), Expect(2) = 1e-65
Identities = 20/24 (83%), Positives = 22/24 (91%)
Frame = -3
Query: 376 RGKAVKALKTHNGDIVGAIMELTT 305
R KAVKALK+H+GDIV AIMELTT
Sbjct: 194 RSKAVKALKSHDGDIVSAIMELTT 217
>gb|AAK48972.1|AF370545_1 alpha NAC-like protein [Arabidopsis thaliana]
gi|18377570|gb|AAL66951.1| alpha NAC-like protein
[Arabidopsis thaliana]
Length = 217
Score = 230 bits (586), Expect(2) = 3e-65
Identities = 131/208 (62%), Positives = 156/208 (74%), Gaps = 2/208 (0%)
Frame = -2
Query: 986 MAPGP-VVDASSEAEALEQVLSAEETTLKKKPQPQEDDAPMVEDVKDDDKDEADDDEDED 810
M+P P VV S++ + + ++A L+KK Q E P+VEDVKDD+ D+ DD+E+ED
Sbjct: 1 MSPPPAVVTESADGQPEQPPVTAIAEELEKKLQTDE---PIVEDVKDDEDDDDDDEEEED 57
Query: 809 DDDEDDDKEDGALGGSEGSKQSRSEKKSRKARLKLGLKPVTGVSRVTIKRTKNILFSISK 630
DD A G S SKQSRSEKKSRKA LKLG+KPVTGVSRVTIKRTKN+LF ISK
Sbjct: 58 DD---------AQGVSGSSKQSRSEKKSRKAMLKLGMKPVTGVSRVTIKRTKNVLFFISK 108
Query: 629 PDVFKSPNSETYVICGEAKIEDLSSQLQTQAAQQFRMPDMGSVMAKQDQGTAADGGQPEE 450
PDVFKSP+SETYVI GEAKIEDLSSQLQTQAAQQF+MP++G+ + + TA Q EE
Sbjct: 109 PDVFKSPHSETYVIFGEAKIEDLSSQLQTQAAQQFKMPEIGATSQRAEASTATVEAQVEE 168
Query: 449 -EEEVDETGVEPHDIDLVMTQAGVSQGQ 369
EEE+DETGVE DIDLVMTQAGVS+ +
Sbjct: 169 DEEEIDETGVEARDIDLVMTQAGVSRSK 196
Score = 42.4 bits (98), Expect(2) = 3e-65
Identities = 20/24 (83%), Positives = 22/24 (91%)
Frame = -3
Query: 376 RGKAVKALKTHNGDIVGAIMELTT 305
R KAVKALK+H+GDIV AIMELTT
Sbjct: 194 RSKAVKALKSHDGDIVSAIMELTT 217
>ref|NP_187845.1| nascent polypeptide associated complex alpha chain, putative;
protein id: At3g12390.1 [Arabidopsis thaliana]
gi|12321974|gb|AAG51031.1|AC069474_30 nascent
polypeptide associated complex alpha chain, putative;
85450-84199 [Arabidopsis thaliana]
gi|15081680|gb|AAK82495.1| AT3g12390/T2E22_130
[Arabidopsis thaliana] gi|15795158|dbj|BAB03146.1|
contains similarity to alpha NAC~gene_id:MQC3.21
[Arabidopsis thaliana] gi|20334844|gb|AAM16178.1|
AT3g12390/T2E22_130 [Arabidopsis thaliana]
Length = 203
Score = 208 bits (529), Expect(2) = 1e-56
Identities = 112/182 (61%), Positives = 142/182 (77%)
Frame = -2
Query: 920 EETTLKKKPQPQEDDAPMVEDVKDDDKDEADDDEDEDDDDEDDDKEDGALGGSEGSKQSR 741
E+ L K + Q+ D E V+DDD +E DD +D+D DD++ D DG GG SKQSR
Sbjct: 5 EKEILAAKLEEQKIDLDKPE-VEDDDDNEDDDSDDDDKDDDEADGLDGEAGGK--SKQSR 61
Query: 740 SEKKSRKARLKLGLKPVTGVSRVTIKRTKNILFSISKPDVFKSPNSETYVICGEAKIEDL 561
SEKKSRKA LKLG+KP+TGVSRVT+K++KNILF ISKPDVFKSP S+TYVI GEAKIEDL
Sbjct: 62 SEKKSRKAMLKLGMKPITGVSRVTVKKSKNILFVISKPDVFKSPASDTYVIFGEAKIEDL 121
Query: 560 SSQLQTQAAQQFRMPDMGSVMAKQDQGTAADGGQPEEEEEVDETGVEPHDIDLVMTQAGV 381
SSQ+Q+QAA+QF+ PD+ +V++K + +AA +++EEVDE GVEP DI+LVMTQAGV
Sbjct: 122 SSQIQSQAAEQFKAPDLSNVISKGESSSAA---VVQDDEEVDEEGVEPKDIELVMTQAGV 178
Query: 380 SQ 375
S+
Sbjct: 179 SR 180
Score = 35.4 bits (80), Expect(2) = 1e-56
Identities = 18/24 (75%), Positives = 19/24 (79%)
Frame = -3
Query: 376 RGKAVKALKTHNGDIVGAIMELTT 305
R AVKALK +GDIV AIMELTT
Sbjct: 180 RPNAVKALKAADGDIVSAIMELTT 203
>dbj|BAB89723.1| putative nascent polypeptide associated complex alpha chain [Oryza
sativa (japonica cultivar-group)]
gi|20161322|dbj|BAB90246.1| putative nascent polypeptide
associated complex alpha chain [Oryza sativa (japonica
cultivar-group)]
Length = 202
Score = 207 bits (527), Expect(2) = 1e-56
Identities = 114/186 (61%), Positives = 143/186 (76%), Gaps = 2/186 (1%)
Frame = -2
Query: 920 EETTLKKKPQPQ--EDDAPMVEDVKDDDKDEADDDEDEDDDDEDDDKEDGALGGSEGSKQ 747
+E L+K + Q E D P++ED DDD D+D+D+D+DD+DDD E S SKQ
Sbjct: 7 QEELLRKHLEEQKIEGDEPILED--DDD----DEDDDDDEDDKDDDVEGAGGDASGRSKQ 60
Query: 746 SRSEKKSRKARLKLGLKPVTGVSRVTIKRTKNILFSISKPDVFKSPNSETYVICGEAKIE 567
SRSEKKSRKA KLG+K +TGVSRVTIK++KNILF ISKPDVFKSPNS+TYVI GEAKIE
Sbjct: 61 SRSEKKSRKAMQKLGMKTITGVSRVTIKKSKNILFVISKPDVFKSPNSDTYVIFGEAKIE 120
Query: 566 DLSSQLQTQAAQQFRMPDMGSVMAKQDQGTAADGGQPEEEEEVDETGVEPHDIDLVMTQA 387
DLSSQLQTQAA+QF+ PD+ +V++K + AA +++EEVDE+GVEP DI+LVMTQA
Sbjct: 121 DLSSQLQTQAAEQFKAPDLSNVISKAEPSAAA-----QDDEEVDESGVEPKDIELVMTQA 175
Query: 386 GVSQGQ 369
VS+ +
Sbjct: 176 TVSRSR 181
Score = 36.2 bits (82), Expect(2) = 1e-56
Identities = 18/23 (78%), Positives = 19/23 (82%)
Frame = -3
Query: 376 RGKAVKALKTHNGDIVGAIMELT 308
R +AVKALK NGDIV AIMELT
Sbjct: 179 RSRAVKALKAANGDIVTAIMELT 201
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 961,543,884
Number of Sequences: 1393205
Number of extensions: 23776713
Number of successful extensions: 455629
Number of sequences better than 10.0: 6046
Number of HSP's better than 10.0 without gapping: 122685
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 264753
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 65119911172
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)