Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC013299A_C01 KMC013299A_c01
(882 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
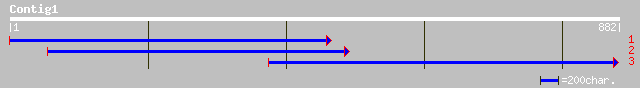
dbj|BAC15989.1| similar to auxin-responsive protein [Oryza sativ... 159 7e-38
ref|NP_174134.1| auxin-regulated GH3 protein, putative; protein ... 139 4e-32
ref|NP_182296.1| putative auxin-responsive protein; protein id: ... 134 2e-30
ref|NP_196840.1| auxin reponsive - like protein; protein id: At5... 133 4e-30
ref|NP_173729.1| GH3-like auxin-regulated protein; protein id: A... 132 9e-30
>dbj|BAC15989.1| similar to auxin-responsive protein [Oryza sativa (japonica
cultivar-group)]
Length = 588
Score = 159 bits (401), Expect = 7e-38
Identities = 83/138 (60%), Positives = 99/138 (71%), Gaps = 1/138 (0%)
Frame = -2
Query: 881 KLLEPQNVLLVDYTSYPDTCLAPGHYVLYWEILHQNTKKTESGASLDTNVLQECCIAVEE 702
K+LE QN LL++YTSY D PGHYVL+WEI + E A LD +L+ CC AVEE
Sbjct: 453 KILENQNYLLLEYTSYTDISTVPGHYVLFWEI---KSTHDERPAPLDAQLLESCCAAVEE 509
Query: 701 QLDYVYCHLR-HDGSIGPLEIRVVEAGTFDTLLDLFISLGASIHQYQTPRGIKSDKALNL 525
LDYVY R HD SIGPLEIR+VEAG FD L+DL +S G+SI+QY+TPR I+S AL L
Sbjct: 510 SLDYVYRRCRAHDRSIGPLEIRLVEAGAFDALMDLLVSHGSSINQYKTPRCIESSLALKL 569
Query: 524 LNSKVTASFFSPRYPQWG 471
LNSKV A FFSP+ P+ G
Sbjct: 570 LNSKVIACFFSPQDPECG 587
>ref|NP_174134.1| auxin-regulated GH3 protein, putative; protein id: At1g28130.1,
supported by cDNA: gi_15450364 [Arabidopsis thaliana]
gi|25345305|pir||B86407 auxin-regulated protein GH3
homolog F3H9.21 - Arabidopsis thaliana
gi|9795624|gb|AAF98442.1|AC021044_21 Unknown protein
[Arabidopsis thaliana]
gi|12322991|gb|AAG51481.1|AC069471_12 unknown protein
[Arabidopsis thaliana] gi|15450365|gb|AAK96476.1|
At1g28130/F3H9_19 [Arabidopsis thaliana]
gi|21360519|gb|AAM47375.1| At1g28130/F3H9_19
[Arabidopsis thaliana]
Length = 609
Score = 139 bits (351), Expect = 4e-32
Identities = 69/136 (50%), Positives = 92/136 (66%), Gaps = 1/136 (0%)
Frame = -2
Query: 878 LLEPQNVLLVDYTSYPDTCLAPGHYVLYWEILHQNTKKTESGASLDTNVLQECCIAVEEQ 699
L P ++LL +YTSY DT PGHYVL+WE+ + + LD +++CC VE+
Sbjct: 470 LQHPSSLLLTEYTSYADTSSIPGHYVLFWEL---KPRHSNDPPKLDDKTMEDCCSEVEDC 526
Query: 698 LDYVYCHLRH-DGSIGPLEIRVVEAGTFDTLLDLFISLGASIHQYQTPRGIKSDKALNLL 522
LDYVY R+ D SIGPLEIRVV GTFD+L+D +S G+S++QY+TPR +KS AL +L
Sbjct: 527 LDYVYRRCRNRDKSIGPLEIRVVSLGTFDSLMDFCVSQGSSLNQYKTPRCVKSGGALEIL 586
Query: 521 NSKVTASFFSPRYPQW 474
+S+V FFS R PQW
Sbjct: 587 DSRVIGRFFSKRVPQW 602
>ref|NP_182296.1| putative auxin-responsive protein; protein id: At2g47750.1,
supported by cDNA: gi_15810039 [Arabidopsis thaliana]
gi|25345300|pir||A84919 auxin-regulated protein GH3
homolog At2g47750 - Arabidopsis thaliana
gi|3738288|gb|AAC63630.1| putative auxin-responsive
protein [Arabidopsis thaliana]
gi|15810040|gb|AAL06947.1| At2g47750/F17A22.14
[Arabidopsis thaliana] gi|23308451|gb|AAN18195.1|
At2g47750/F17A22.14 [Arabidopsis thaliana]
Length = 585
Score = 134 bits (336), Expect = 2e-30
Identities = 70/132 (53%), Positives = 87/132 (65%), Gaps = 2/132 (1%)
Frame = -2
Query: 863 NVLLVDYTSYPDTCLAPGHYVLYWEILHQNTKKTESGASLDTNVLQECCIAVEEQLDYVY 684
N L +YTSY DT PGHYVL+WEI L+ +++ECC+AVEE+LDY+Y
Sbjct: 462 NAFLAEYTSYADTSSVPGHYVLFWEIQ----------GHLEPKLMEECCVAVEEELDYIY 511
Query: 683 CHLR-HDGSIGPLEIRVVEAGTFDTLLDLFISLGASIHQYQTPRGIKSDKA-LNLLNSKV 510
R + SIG LEIRVV+ GTF+ L+DL IS G S +QY+TPR +KS+ A LLN V
Sbjct: 512 RQCRTKERSIGALEIRVVKPGTFEKLMDLIISQGGSFNQYKTPRCVKSNSATFKLLNGHV 571
Query: 509 TASFFSPRYPQW 474
ASFFSPR P W
Sbjct: 572 MASFFSPRDPTW 583
>ref|NP_196840.1| auxin reponsive - like protein; protein id: At5g13360.1
[Arabidopsis thaliana] gi|11281349|pir||T48583
auxin-regulated protein GH3 homolog T22N19.10 -
Arabidopsis thaliana gi|7543903|emb|CAB87143.1| auxin
reponsive-like protein [Arabidopsis thaliana]
Length = 594
Score = 133 bits (334), Expect = 4e-30
Identities = 64/136 (47%), Positives = 93/136 (68%), Gaps = 1/136 (0%)
Frame = -2
Query: 878 LLEPQNVLLVDYTSYPDTCLAPGHYVLYWEILHQNTKKTESGASLDTNVLQECCIAVEEQ 699
LLEP +++L+D+TS D+ PGHYVLYWE+ K ++ LD NVL+ECC +EE
Sbjct: 453 LLEPHDLMLMDFTSRVDSSSYPGHYVLYWEL---GRKVKDAKLELDQNVLEECCFTIEES 509
Query: 698 LDYVYCHLR-HDGSIGPLEIRVVEAGTFDTLLDLFISLGASIHQYQTPRGIKSDKALNLL 522
LD VY R +D +IGPLEI+VV+ G FD L++ F+S G+S+ QY+TPR + +++AL +L
Sbjct: 510 LDAVYRKGRKNDKNIGPLEIKVVKPGAFDKLMNFFLSRGSSVSQYKTPRSVTNEEALKIL 569
Query: 521 NSKVTASFFSPRYPQW 474
+ V + F S + P W
Sbjct: 570 EANVVSEFLSQKTPSW 585
>ref|NP_173729.1| GH3-like auxin-regulated protein; protein id: At1g23160.1
[Arabidopsis thaliana] gi|25345307|pir||H86365 T26J12.7
protein - Arabidopsis thaliana gi|2829896|gb|AAC00604.1|
highly similar to auxin-regulated protein GH3,
gp|X60033|18591 [Arabidopsis thaliana]
Length = 578
Score = 132 bits (331), Expect = 9e-30
Identities = 66/131 (50%), Positives = 90/131 (68%), Gaps = 3/131 (2%)
Frame = -2
Query: 875 LEPQNVLLVDYTSYPDTCLAPGHYVLYWEILHQNT-KKTESGASLDTNVLQECCIAVEEQ 699
L+ +++L+D+TSY D PGHYV+YWE+ ++N KK++ L ECC+ +E+
Sbjct: 448 LDSSHLMLIDFTSYADISTIPGHYVVYWEVKNKNEDKKSKKHIELKEETFSECCLLMEDS 507
Query: 698 LDYVY--CHLRHDGSIGPLEIRVVEAGTFDTLLDLFISLGASIHQYQTPRGIKSDKALNL 525
LD VY C + + S+GPLEI+VV GTFD+L+D FIS GASI QY+TPR IKS KAL +
Sbjct: 508 LDSVYKICRFKEE-SVGPLEIKVVRQGTFDSLMDYFISQGASIGQYKTPRCIKSGKALEV 566
Query: 524 LNSKVTASFFS 492
L V A+FFS
Sbjct: 567 LEENVVATFFS 577
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 754,620,447
Number of Sequences: 1393205
Number of extensions: 16685692
Number of successful extensions: 44752
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 42614
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44673
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 47660818527
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)