Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC013068A_C02 KMC013068A_c02
(1578 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
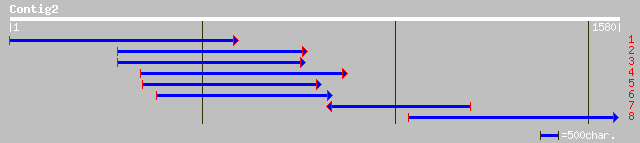
ref|NP_181435.1| unknown protein; protein id: At2g39020.1 [Arabi... 306 6e-82
ref|NP_565898.1| expressed protein; protein id: At2g39030.1, sup... 286 8e-76
gb|AAK96852.1| Unknown protein [Arabidopsis thaliana] gi|1837743... 271 3e-71
dbj|BAC23029.1| tyramine hydroxycinnamoyl transferase [Solanum t... 162 1e-38
gb|AAL99189.1| N-hydroxycinnamoyl-CoA:tyramine N-hydroxycinnamoy... 161 2e-38
>ref|NP_181435.1| unknown protein; protein id: At2g39020.1 [Arabidopsis thaliana]
gi|25408632|pir||B84812 hypothetical protein At2g39020
[imported] - Arabidopsis thaliana
gi|3928086|gb|AAC79612.1| unknown protein [Arabidopsis
thaliana] gi|28466877|gb|AAO44047.1| At2g39020
[Arabidopsis thaliana]
Length = 236
Score = 306 bits (784), Expect = 6e-82
Identities = 156/233 (66%), Positives = 184/233 (78%), Gaps = 4/233 (1%)
Frame = -3
Query: 1213 MAAAAPPPAPTPAATS---LPETATHRHPVFTRIRLAVPSDVPHIHKMTYQMAVFERLTH 1043
MAAAAPPP PT A P + HP+F+RIRLA PSDVP IHK+ +QMAVFERLTH
Sbjct: 1 MAAAAPPPPPTAAPEPNMVAPLISPIGHPMFSRIRLATPSDVPFIHKLIHQMAVFERLTH 60
Query: 1042 LFATTESSLTSTLFSPDNKPFHSFTVFILEVSSNPFTDTHFDN-DPFYKPVTKTVHLELP 866
LF+ TES L STLF+ ++PF SFTVF+LEVS +PF T + P + P KT +L+LP
Sbjct: 61 LFSATESGLASTLFT--SRPFQSFTVFLLEVSRSPFPATITSSPSPDFTPFFKTHNLDLP 118
Query: 865 LDDPEKETFRNQLGNEVFVAGFVLFSPNYSTFLAKPGFYVEDLFVRECYRRKGFGRMLLS 686
+DDPE F + N+V VAGFVLF PNYS+FL+KPGFY+ED+FVRE YRRKGFG MLL+
Sbjct: 119 IDDPESYNFSPDMLNDVVVAGFVLFFPNYSSFLSKPGFYIEDIFVREPYRRKGFGSMLLT 178
Query: 685 AVAKQAVKMGYGRVEWVVLDWNVNAIRFYEDMGAKILQEWRLCRLTGDDLQAY 527
AVAKQAVKMGYGRVEWVVLDWNVNAI+FYE MGA+ILQEWR+CRLTGD L+A+
Sbjct: 179 AVAKQAVKMGYGRVEWVVLDWNVNAIKFYEQMGAQILQEWRVCRLTGDALEAF 231
>ref|NP_565898.1| expressed protein; protein id: At2g39030.1, supported by cDNA:
gi_15451161 [Arabidopsis thaliana]
gi|25408633|pir||C84812 hypothetical protein At2g39030
[imported] - Arabidopsis thaliana
gi|3928087|gb|AAC79613.1| expressed protein [Arabidopsis
thaliana]
Length = 228
Score = 286 bits (731), Expect = 8e-76
Identities = 135/222 (60%), Positives = 173/222 (77%)
Frame = -3
Query: 1195 PPAPTPAATSLPETATHRHPVFTRIRLAVPSDVPHIHKMTYQMAVFERLTHLFATTESSL 1016
PP P ++PET+ H +F+RIRLA P+DVP IHK+ +QMAVFERLTHLF TES L
Sbjct: 3 PPTAAPEPNTVPETSPTGHRMFSRIRLATPTDVPFIHKLIHQMAVFERLTHLFVATESGL 62
Query: 1015 TSTLFSPDNKPFHSFTVFILEVSSNPFTDTHFDNDPFYKPVTKTVHLELPLDDPEKETFR 836
STLF+ ++PF + TVF+LE+S +PF TH + P + P +T ++LP++DP++E F
Sbjct: 63 ASTLFN--SRPFQAVTVFLLEISPSPFPTTHDASSPDFTPFLETHKVDLPIEDPDREKFL 120
Query: 835 NQLGNEVFVAGFVLFSPNYSTFLAKPGFYVEDLFVRECYRRKGFGRMLLSAVAKQAVKMG 656
N+V VAGFVLF PNY +FLAK GFY+ED+F+RE YRRKGFG++LL+AVAKQAVK+G
Sbjct: 121 PDKLNDVVVAGFVLFFPNYPSFLAKQGFYIEDIFMREPYRRKGFGKLLLTAVAKQAVKLG 180
Query: 655 YGRVEWVVLDWNVNAIRFYEDMGAKILQEWRLCRLTGDDLQA 530
GRVEW+V+DWNVNAI FYE MGA++ +EWRLCRLTGD LQA
Sbjct: 181 VGRVEWIVIDWNVNAINFYEQMGAQVFKEWRLCRLTGDALQA 222
>gb|AAK96852.1| Unknown protein [Arabidopsis thaliana] gi|18377432|gb|AAL66882.1|
unknown protein [Arabidopsis thaliana]
Length = 206
Score = 271 bits (692), Expect = 3e-71
Identities = 128/202 (63%), Positives = 163/202 (80%)
Frame = -3
Query: 1135 VFTRIRLAVPSDVPHIHKMTYQMAVFERLTHLFATTESSLTSTLFSPDNKPFHSFTVFIL 956
+F+RIRLA P+DVP IHK+ +QMAVFERLTHLF TES L STLF+ ++PF + TVF+L
Sbjct: 1 MFSRIRLATPTDVPFIHKLIHQMAVFERLTHLFVATESGLASTLFN--SRPFQAVTVFLL 58
Query: 955 EVSSNPFTDTHFDNDPFYKPVTKTVHLELPLDDPEKETFRNQLGNEVFVAGFVLFSPNYS 776
E+S +PF TH + P + P +T ++LP++DP++E F N+V VAGFVLF PNY
Sbjct: 59 EISPSPFPTTHDASSPDFTPFLETHKVDLPIEDPDREKFLPDKLNDVVVAGFVLFFPNYP 118
Query: 775 TFLAKPGFYVEDLFVRECYRRKGFGRMLLSAVAKQAVKMGYGRVEWVVLDWNVNAIRFYE 596
+FLAK GFY+ED+F+RE YRRKGFG++LL+AVAKQAVK+G GRVEW+V+DWNVNAI FYE
Sbjct: 119 SFLAKQGFYIEDIFMREPYRRKGFGKLLLTAVAKQAVKLGVGRVEWIVIDWNVNAINFYE 178
Query: 595 DMGAKILQEWRLCRLTGDDLQA 530
MGA++ +EWRLCRLTGD LQA
Sbjct: 179 QMGAQVFKEWRLCRLTGDALQA 200
>dbj|BAC23029.1| tyramine hydroxycinnamoyl transferase [Solanum tuberosum]
Length = 247
Score = 162 bits (410), Expect = 1e-38
Identities = 87/235 (37%), Positives = 134/235 (57%), Gaps = 9/235 (3%)
Frame = -3
Query: 1204 AAPPPAPTPAATSLPETATHRHPV------FTRIRLAVPSDVPHIHKMTYQMAVFERLTH 1043
A P PTP+ T + + ++ + V +TR+RLA SD+ HI+++ YQ+ V+ TH
Sbjct: 2 APAPQQPTPSETIITDASSENNNVTITGKIYTRVRLATKSDLSHIYQLFYQIHVYHNFTH 61
Query: 1042 LFATTESSLTSTLFSPDNKP-FHSFTVFILEVSSNPFTDTHFDNDPFYKPVTKTVHLELP 866
L+ TESSL LF + P F+ +V +LEVS PF + D + PV T L+ P
Sbjct: 62 LYKATESSLEGLLFKENPLPLFYGPSVLLLEVSPTPFNEPKNTTDEGFNPVLTTFDLKFP 121
Query: 865 LDDPEKETFRNQLGN--EVFVAGFVLFSPNYSTFLAKPGFYVEDLFVRECYRRKGFGRML 692
+ + + E FR++ + + ++AG+ F NYS F KPGFY E L+ RE YR+ G G++L
Sbjct: 122 VVEGQVEEFRSKYDDKSDAYIAGYAFFYANYSCFNDKPGFYFESLYFRESYRKLGMGKLL 181
Query: 691 LSAVAKQAVKMGYGRVEWVVLDWNVNAIRFYEDMGAKILQEWRLCRLTGDDLQAY 527
V+ A G+ V+ +V WN + FY +MG +I E+R +L G++LQ Y
Sbjct: 182 FGTVSSIAADNGFVSVDGIVAVWNKKSYDFYINMGVEIFDEFRYGKLHGENLQKY 236
>gb|AAL99189.1| N-hydroxycinnamoyl-CoA:tyramine N-hydroxycinnamoyl transferase THT1-3
[Lycopersicon esculentum]
Length = 231
Score = 161 bits (408), Expect = 2e-38
Identities = 90/226 (39%), Positives = 129/226 (56%), Gaps = 4/226 (1%)
Frame = -3
Query: 1192 PAPTPAATS-LPETATHRHPVFTRIRLAVPSDVPHIHKMTYQMAVFERLTHLFATTESSL 1016
PA A TS T ++TR+RLA SD+ HI+++ YQ+ + THL+ TESSL
Sbjct: 3 PALEQAITSDASSDVTITGKIYTRVRLATKSDLSHIYRLFYQIHEYHNYTHLYKATESSL 62
Query: 1015 TSTLFSPDNKP-FHSFTVFILEVSSNPFTDTHFDNDPFYKPVTKTVHLELPLDDPEKETF 839
+ LF + P F+ +V +LEVS PF + D +KPV T L+ P+ + E E F
Sbjct: 63 ANLLFKENPLPLFYGPSVLLLEVSPTPFDEPKNTTDEGFKPVLTTFDLKFPVVEGEVEEF 122
Query: 838 RNQLGN--EVFVAGFVLFSPNYSTFLAKPGFYVEDLFVRECYRRKGFGRMLLSAVAKQAV 665
R++ + +V++AG+ F NYS F KPGFY E L+ RE YR+ G G +L VA A
Sbjct: 123 RSKYDDKSDVYIAGYAFFYANYSCFYDKPGFYFESLYFRESYRKLGMGSLLFGTVASIAA 182
Query: 664 KMGYGRVEWVVLDWNVNAIRFYEDMGAKILQEWRLCRLTGDDLQAY 527
G+ VE +V WN + FY +MG +I E+R +L G++LQ Y
Sbjct: 183 NNGFVSVEGIVAVWNKKSYDFYVNMGVEIFDEFRYGKLHGENLQKY 228
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,532,358,972
Number of Sequences: 1393205
Number of extensions: 42960831
Number of successful extensions: 510302
Number of sequences better than 10.0: 6695
Number of HSP's better than 10.0 without gapping: 181930
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 370500
length of database: 448,689,247
effective HSP length: 128
effective length of database: 270,359,007
effective search space used: 107332525779
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)