Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
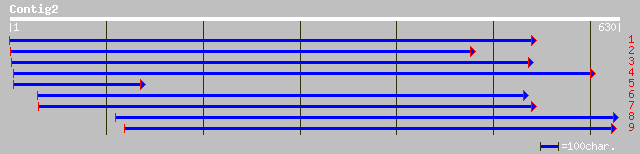
Query= KMC012987A_C02 KMC012987A_c02
(630 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO00883.1| threonine dehydratase/deaminase (OMR1) [Arabidops... 179 2e-44
ref|NP_187616.1| threonine dehydratase/deaminase (OMR1); protein... 179 2e-44
gb|AAD54324.1|AF177212_1 threonine dehydratase/deaminase [Arabid... 174 7e-43
gb|AAL58211.1|AC090882_14 putative dehydratase/deaminase [Oryza ... 158 6e-38
gb|AAG59585.1|AF229927_1 threonine deaminase [Nicotiana attenuata] 149 3e-35
>gb|AAO00883.1| threonine dehydratase/deaminase (OMR1) [Arabidopsis thaliana]
Length = 592
Score = 179 bits (455), Expect = 2e-44
Identities = 84/119 (70%), Positives = 103/119 (85%)
Frame = -2
Query: 629 MKEKLESSQLETHDLTESELVKDHLRYLMGGRSNVQNEVLCRFTFPERPGALMNFLDSFS 450
+++++ESSQL+T +LT S+LVKDHLRYLMGGRS V +EVLCRFTFPERPGALMNFLDSFS
Sbjct: 474 LQKRMESSQLKTVNLTTSDLVKDHLRYLMGGRSTVGDEVLCRFTFPERPGALMNFLDSFS 533
Query: 449 PRWNISLFHYRAEGAARADILVGIQVPRNEMDEFHGRANRLGYEYKVVNNDDDFQLLMH 273
PRWNI+LFHYR +G A++LVGIQVP EM+EF RA LGY+Y +V++DD F+LLMH
Sbjct: 534 PRWNITLFHYRGQGETGANVLVGIQVPEQEMEEFKNRAKALGYDYFLVSDDDYFKLLMH 592
>ref|NP_187616.1| threonine dehydratase/deaminase (OMR1); protein id: At3g10050.1,
supported by cDNA: gi_18086438, supported by cDNA:
gi_3800877, supported by cDNA: gi_5853118 [Arabidopsis
thaliana] gi|17369978|sp|Q9ZSS6|THD1_ARATH Threonine
dehydratase biosynthetic, chloroplast precursor
(Threonine deaminase) (TD) gi|11358880|pir||T51712
threonine ammonia-lyase (EC 4.3.1.19) [similarity] -
Arabidopsis thaliana gi|3800878|gb|AAC97936.1| threonine
dehydratase/deaminase [Arabidopsis thaliana]
gi|6143871|gb|AAF04418.1|AC010927_11 threonine
dehydratase/deaminase (OMR1) [Arabidopsis thaliana]
gi|6942237|gb|AAF32370.1|AF221984_1 threonine
dehydratase/deaminase [Arabidopsis thaliana]
gi|18086439|gb|AAL57674.1| AT3g10050/T22K18_12
[Arabidopsis thaliana]
Length = 592
Score = 179 bits (455), Expect = 2e-44
Identities = 84/119 (70%), Positives = 103/119 (85%)
Frame = -2
Query: 629 MKEKLESSQLETHDLTESELVKDHLRYLMGGRSNVQNEVLCRFTFPERPGALMNFLDSFS 450
+++++ESSQL+T +LT S+LVKDHLRYLMGGRS V +EVLCRFTFPERPGALMNFLDSFS
Sbjct: 474 LQKRMESSQLKTVNLTTSDLVKDHLRYLMGGRSTVGDEVLCRFTFPERPGALMNFLDSFS 533
Query: 449 PRWNISLFHYRAEGAARADILVGIQVPRNEMDEFHGRANRLGYEYKVVNNDDDFQLLMH 273
PRWNI+LFHYR +G A++LVGIQVP EM+EF RA LGY+Y +V++DD F+LLMH
Sbjct: 534 PRWNITLFHYRGQGETGANVLVGIQVPEQEMEEFKNRAKALGYDYFLVSDDDYFKLLMH 592
>gb|AAD54324.1|AF177212_1 threonine dehydratase/deaminase [Arabidopsis thaliana]
Length = 592
Score = 174 bits (442), Expect = 7e-43
Identities = 82/119 (68%), Positives = 101/119 (83%)
Frame = -2
Query: 629 MKEKLESSQLETHDLTESELVKDHLRYLMGGRSNVQNEVLCRFTFPERPGALMNFLDSFS 450
+++++ESSQL+T +LT S+LVKDHL YLMGGRS V +EVLCRFTFPERPGALMNFLDSFS
Sbjct: 474 LQKRMESSQLKTVNLTTSDLVKDHLCYLMGGRSTVGDEVLCRFTFPERPGALMNFLDSFS 533
Query: 449 PRWNISLFHYRAEGAARADILVGIQVPRNEMDEFHGRANRLGYEYKVVNNDDDFQLLMH 273
PRWNI+LFHY +G A++LVGIQVP EM+EF RA LGY+Y +V++DD F+LLMH
Sbjct: 534 PRWNITLFHYHGQGETGANVLVGIQVPEQEMEEFKNRAKALGYDYFLVSDDDYFKLLMH 592
>gb|AAL58211.1|AC090882_14 putative dehydratase/deaminase [Oryza sativa (japonica
cultivar-group)]
Length = 602
Score = 158 bits (399), Expect = 6e-38
Identities = 74/118 (62%), Positives = 94/118 (78%)
Frame = -2
Query: 629 MKEKLESSQLETHDLTESELVKDHLRYLMGGRSNVQNEVLCRFTFPERPGALMNFLDSFS 450
M E++ESS+L T DLT+++L KDHLRY +GGRS V +E++ RF FPERPGALM FLD+FS
Sbjct: 480 MVERMESSKLRTVDLTDNDLAKDHLRYFIGGRSEVTDELVYRFIFPERPGALMKFLDAFS 539
Query: 449 PRWNISLFHYRAEGAARADILVGIQVPRNEMDEFHGRANRLGYEYKVVNNDDDFQLLM 276
PRWNISLFHYRA+G A++LVGIQVP E DEF RA+ LGYEY N++ ++LL+
Sbjct: 540 PRWNISLFHYRAQGETGANVLVGIQVPPEEFDEFKSRADNLGYEYMSEQNNEIYRLLL 597
>gb|AAG59585.1|AF229927_1 threonine deaminase [Nicotiana attenuata]
Length = 601
Score = 149 bits (376), Expect = 3e-35
Identities = 68/118 (57%), Positives = 91/118 (76%)
Frame = -2
Query: 629 MKEKLESSQLETHDLTESELVKDHLRYLMGGRSNVQNEVLCRFTFPERPGALMNFLDSFS 450
M E+++SSQL T +LT + LVK+HLR+LMGGRS NE+ C+F FPE+PGAL FLD+FS
Sbjct: 483 MLERMKSSQLNTVNLTNNNLVKEHLRHLMGGRSEPSNEIFCQFIFPEKPGALRKFLDAFS 542
Query: 449 PRWNISLFHYRAEGAARADILVGIQVPRNEMDEFHGRANRLGYEYKVVNNDDDFQLLM 276
PRWNISLFHYR +G A +LVG QVP+ E++EF +AN LGY Y++ + ++ QL+M
Sbjct: 543 PRWNISLFHYREQGELDASVLVGFQVPKGEIEEFRVQANNLGYSYEIESLNEASQLIM 600
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 524,888,413
Number of Sequences: 1393205
Number of extensions: 11427978
Number of successful extensions: 26072
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 25271
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26007
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25870486187
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)