Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC012944A_C01 KMC012944A_c01
(545 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
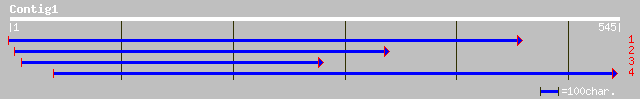
ref|NP_190059.2| kinesin-related protein; protein id: At3g44730.... 44 0.001
gb|AAK92458.3|AF398149_1 kinesin-like protein heavy chain [Arabi... 44 0.001
gb|ZP_00076987.1| hypothetical protein [Methanosarcina barkeri] 34 1.3
ref|XP_218821.1| similar to AT11276p [Drosophila melanogaster] [... 32 6.5
ref|ZP_00060137.1| hypothetical protein [Clostridium thermocellu... 32 6.5
>ref|NP_190059.2| kinesin-related protein; protein id: At3g44730.1 [Arabidopsis
thaliana]
Length = 1089
Score = 44.3 bits (103), Expect = 0.001
Identities = 37/95 (38%), Positives = 49/95 (50%), Gaps = 1/95 (1%)
Frame = -2
Query: 493 DPSPKSDYSEHENELRLVESAVHSAINIKKIHQNLPKNFQNLESRGIVQTGEPILATKVE 314
D + KSD SE +NE K +N KN N + R I + L V+
Sbjct: 1006 DSTLKSDSSETDNE---------PPSKSKNAQRNSSKNSLNHKLRTIYAHEDTSL---VD 1053
Query: 313 NKLFNGSGSNIKEGTN-ASMPEFRRSRSTPRGKFL 212
+K NG+ ++IKEG N SMPEFRRSRST +F+
Sbjct: 1054 DKPSNGT-AHIKEGNNNISMPEFRRSRSTHHARFM 1087
>gb|AAK92458.3|AF398149_1 kinesin-like protein heavy chain [Arabidopsis thaliana]
Length = 1087
Score = 44.3 bits (103), Expect = 0.001
Identities = 37/95 (38%), Positives = 49/95 (50%), Gaps = 1/95 (1%)
Frame = -2
Query: 493 DPSPKSDYSEHENELRLVESAVHSAINIKKIHQNLPKNFQNLESRGIVQTGEPILATKVE 314
D + KSD SE +NE K +N KN N + R I + L V+
Sbjct: 1004 DSTLKSDSSETDNE---------PPSKSKNAQRNSSKNSLNHKLRTIYAHEDTSL---VD 1051
Query: 313 NKLFNGSGSNIKEGTN-ASMPEFRRSRSTPRGKFL 212
+K NG+ ++IKEG N SMPEFRRSRST +F+
Sbjct: 1052 DKPSNGT-AHIKEGNNNISMPEFRRSRSTHHARFM 1085
>gb|ZP_00076987.1| hypothetical protein [Methanosarcina barkeri]
Length = 1066
Score = 33.9 bits (76), Expect = 1.3
Identities = 30/101 (29%), Positives = 46/101 (44%), Gaps = 7/101 (6%)
Frame = -2
Query: 511 TGEMTVDPSPKSDYSEHENELRL-VESAVHSAINIKKIHQNLPKNFQNLESRGIVQTGEP 335
T E+ + + D + NEL + VE AV A+N + L + F E ++Q E
Sbjct: 514 TLEVEAELQRRQDERDLLNELHIQVEKAVDKALNDNDLE--LEEYFNQKEKLKVLQEHEY 571
Query: 334 ILATKVENKLFNGSGSNIKEGTN------ASMPEFRRSRST 230
L TK++ L N + NIK G A +P+ +R T
Sbjct: 572 DLKTKIDAFLCNLNEGNIKSGDKDSWLLLAGVPDLQRELET 612
>ref|XP_218821.1| similar to AT11276p [Drosophila melanogaster] [Rattus norvegicus]
Length = 866
Score = 31.6 bits (70), Expect = 6.5
Identities = 18/56 (32%), Positives = 31/56 (55%), Gaps = 4/56 (7%)
Frame = -2
Query: 544 HHQLSPYK-IQFTGEMTVDPSPKSDYSEHENELRLVESAV---HSAINIKKIHQNL 389
H +L Y+ + G ++ +P P SD+ E EL+ VESA+ A+ +KI + +
Sbjct: 591 HRKLHEYRGVAGPGSISREPDPDSDWEPEERELQEVESALKRQKKAMRFQKIRRQM 646
>ref|ZP_00060137.1| hypothetical protein [Clostridium thermocellum ATCC 27405]
Length = 3705
Score = 31.6 bits (70), Expect = 6.5
Identities = 20/71 (28%), Positives = 33/71 (46%)
Frame = -2
Query: 472 YSEHENELRLVESAVHSAINIKKIHQNLPKNFQNLESRGIVQTGEPILATKVENKLFNGS 293
YS+H + L + + N + N NF+ + +V +GE + E+ +GS
Sbjct: 203 YSQHSHANYLTDGTLEVKGNFTQKRYNSYDNFKATGNHRVVLSGEELQIVTFESSSGSGS 262
Query: 292 GSNIKEGTNAS 260
NI E TN+S
Sbjct: 263 CINILEITNSS 273
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 436,416,457
Number of Sequences: 1393205
Number of extensions: 9156796
Number of successful extensions: 20721
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 20301
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20716
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18660035355
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)