Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
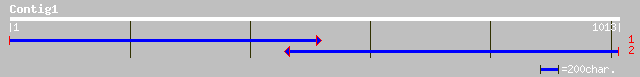
Query= KMC012874A_C01 KMC012874A_c01
(1013 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_568926.1| protein disulfide isomerase precursor - like; p... 324 2e-97
gb|AAM65262.1| protein disulfide isomerase precursor-like [Arabi... 324 2e-97
ref|NP_191056.2| protein disulfide-isomerase-like protein; prote... 277 2e-78
pir||T06724 protein disulfide-isomerase homolog F28P10.60 - Arab... 277 3e-78
gb|AAO26314.1| protein disulphide isomerase [Elaeis guineensis] 233 2e-67
>ref|NP_568926.1| protein disulfide isomerase precursor - like; protein id:
At5g60640.1, supported by cDNA: 3797. [Arabidopsis
thaliana] gi|9759328|dbj|BAB09837.1| protein disulphide
isomerase-like protein [Arabidopsis thaliana]
gi|25082813|gb|AAN72005.1| protein disulfide isomerase
precursor - like [Arabidopsis thaliana]
Length = 597
Score = 324 bits (830), Expect(3) = 2e-97
Identities = 158/268 (58%), Positives = 207/268 (76%), Gaps = 20/268 (7%)
Frame = -1
Query: 959 EEEDLSFLDE---------PDTSSSLHHHDDDGGD------FGDDADLD-DFSGY----E 840
++EDLSFL++ SSS + +GG+ + DD D + DFS
Sbjct: 33 DDEDLSFLEDLKEDDVPGADSLSSSTGFDEFEGGEEEDPDMYNDDDDEEGDFSDLGNPDS 92
Query: 839 DAYKQPEIDDKDVVVFKDKNFSDVVKNNRYVMVEFYAPWCGHCQALAPEYAAAATELKGE 660
D PEID+KDVVV K++NF+DV++NN+YV+VEFYAPWCGHCQ+LAPEYAAAATELK +
Sbjct: 93 DPLPTPEIDEKDVVVIKERNFTDVIENNQYVLVEFYAPWCGHCQSLAPEYAAAATELKED 152
Query: 659 NVILAKVDATEESEXAQEYDIQGFPTLYFFVDGIHKPYNGQRNKEAILTWIKKKTGPGIH 480
V+LAK+DATEE+E AQEY +QGFPTL FFVDG HKPY G R KE I+TW+KKK GPG++
Sbjct: 153 GVVLAKIDATEENELAQEYRVQGFPTLLFFVDGEHKPYTGGRTKETIVTWVKKKIGPGVY 212
Query: 479 NITSLEDAERILTSESKLVVGFINSLVGPESEELAAASRLEDDVNFYPTLNPDVAKLFHI 300
N+T+L+DAE++LTS +K+V+G++NSLVG E ++L AAS+ EDDVNFY T+NPDVAK+FH+
Sbjct: 213 NLTTLDDAEKVLTSGNKVVLGYLNSLVGVEHDQLNAASKAEDDVNFYQTVNPDVAKMFHL 272
Query: 299 DPNVKRPALILIQKEEEKLNHSGGQFRK 216
DP KRPAL+L++KEEEK++H G+F K
Sbjct: 273 DPESKRPALVLVKKEEEKISHFDGEFVK 300
Score = 73.2 bits (178), Expect = 6e-12
Identities = 39/109 (35%), Positives = 62/109 (56%), Gaps = 2/109 (1%)
Frame = -1
Query: 881 FGDDADLDDFSGYEDAYKQPEIDDKDVVVFKDKNFSDVV-KNNRYVMVEFYAPWCGHCQA 705
FG+D D + + PE +D+DV + NF ++V +++ V++E YAPWCGHCQA
Sbjct: 417 FGEDFLNDKLKPFYKSDPIPEKNDEDVKIVVGDNFDEIVLDDSKDVLLEVYAPWCGHCQA 476
Query: 704 LAPEYAAAATELKG-ENVILAKVDATEESEXAQEYDIQGFPTLYFFVDG 561
L P Y A L+ +++++ K+D T + +GFPT+ FF G
Sbjct: 477 LEPMYNKLAKHLRSIDSLVITKMDGTTNEH--PKAKAEGFPTILFFPAG 523
Score = 40.0 bits (92), Expect(3) = 2e-97
Identities = 17/29 (58%), Positives = 24/29 (82%)
Frame = -2
Query: 223 FVKSEIADFVSSNQLPLVTIFTRESAPAV 137
FVKS + FVS+N+L LV++FTRE+AP +
Sbjct: 298 FVKSALVSFVSANKLALVSVFTRETAPEI 326
Score = 37.0 bits (84), Expect(3) = 2e-97
Identities = 17/35 (48%), Positives = 25/35 (70%)
Frame = -3
Query: 123 IQKQLLLFATSTDSDKILPFFLEAAKFFSQLILYM 19
I+KQLLLF T +S+K+L F EAAK F ++++
Sbjct: 331 IKKQLLLFVTKNESEKVLTEFQEAAKSFKGKLIFV 365
>gb|AAM65262.1| protein disulfide isomerase precursor-like [Arabidopsis thaliana]
Length = 597
Score = 324 bits (830), Expect(3) = 2e-97
Identities = 158/268 (58%), Positives = 207/268 (76%), Gaps = 20/268 (7%)
Frame = -1
Query: 959 EEEDLSFLDE---------PDTSSSLHHHDDDGGD------FGDDADLD-DFSGY----E 840
++EDLSFL++ SSS + +GG+ + DD D + DFS
Sbjct: 33 DDEDLSFLEDLKEDDVPGADSLSSSTGFDEFEGGEEEDPDMYNDDDDEEGDFSDLGNPDS 92
Query: 839 DAYKQPEIDDKDVVVFKDKNFSDVVKNNRYVMVEFYAPWCGHCQALAPEYAAAATELKGE 660
D PEID+KDVVV K++NF+DV++NN+YV+VEFYAPWCGHCQ+LAPEYAAAATELK +
Sbjct: 93 DPLPTPEIDEKDVVVIKERNFTDVIENNQYVLVEFYAPWCGHCQSLAPEYAAAATELKED 152
Query: 659 NVILAKVDATEESEXAQEYDIQGFPTLYFFVDGIHKPYNGQRNKEAILTWIKKKTGPGIH 480
V+LAK+DATEE+E AQEY +QGFPTL FFVDG HKPY G R KE I+TW+KKK GPG++
Sbjct: 153 GVVLAKIDATEENELAQEYRVQGFPTLLFFVDGEHKPYTGGRTKETIVTWVKKKIGPGVY 212
Query: 479 NITSLEDAERILTSESKLVVGFINSLVGPESEELAAASRLEDDVNFYPTLNPDVAKLFHI 300
N+T+L+DAE++LTS +K+V+G++NSLVG E ++L AAS+ EDDVNFY T+NPDVAK+FH+
Sbjct: 213 NLTTLDDAEKVLTSGNKVVLGYLNSLVGVEHDQLNAASKAEDDVNFYQTVNPDVAKMFHL 272
Query: 299 DPNVKRPALILIQKEEEKLNHSGGQFRK 216
DP KRPAL+L++KEEEK++H G+F K
Sbjct: 273 DPESKRPALVLVKKEEEKISHFDGEFVK 300
Score = 73.2 bits (178), Expect = 6e-12
Identities = 39/109 (35%), Positives = 62/109 (56%), Gaps = 2/109 (1%)
Frame = -1
Query: 881 FGDDADLDDFSGYEDAYKQPEIDDKDVVVFKDKNFSDVV-KNNRYVMVEFYAPWCGHCQA 705
FG+D D + + PE +D+DV + NF ++V +++ V++E YAPWCGHCQA
Sbjct: 417 FGEDFLNDKLKPFYKSDPIPEKNDEDVKIVVGDNFDEIVLDDSKDVLLEVYAPWCGHCQA 476
Query: 704 LAPEYAAAATELKG-ENVILAKVDATEESEXAQEYDIQGFPTLYFFVDG 561
L P Y A L+ +++++ K+D T + +GFPT+ FF G
Sbjct: 477 LEPMYNKLAKHLRSIDSLVITKMDGTTNEH--PKAKAEGFPTILFFPAG 523
Score = 40.0 bits (92), Expect(3) = 2e-97
Identities = 17/29 (58%), Positives = 24/29 (82%)
Frame = -2
Query: 223 FVKSEIADFVSSNQLPLVTIFTRESAPAV 137
FVKS + FVS+N+L LV++FTRE+AP +
Sbjct: 298 FVKSALVSFVSANKLALVSVFTRETAPEI 326
Score = 37.0 bits (84), Expect(3) = 2e-97
Identities = 17/35 (48%), Positives = 25/35 (70%)
Frame = -3
Query: 123 IQKQLLLFATSTDSDKILPFFLEAAKFFSQLILYM 19
I+KQLLLF T +S+K+L F EAAK F ++++
Sbjct: 331 IKKQLLLFVTKNESEKVLTEFQEAAKSFKGKLIFV 365
>ref|NP_191056.2| protein disulfide-isomerase-like protein; protein id: At3g54960.1,
supported by cDNA: gi_20260431 [Arabidopsis thaliana]
gi|18072841|emb|CAC81067.1| ERp72 [Arabidopsis thaliana]
gi|20260432|gb|AAM13114.1| protein
disulfide-isomerase-like protein [Arabidopsis thaliana]
gi|23197928|gb|AAN15491.1| protein
disulfide-isomerase-like protein [Arabidopsis thaliana]
Length = 579
Score = 277 bits (709), Expect(2) = 2e-78
Identities = 144/263 (54%), Positives = 185/263 (69%), Gaps = 16/263 (6%)
Frame = -1
Query: 956 EEDLSFLDEPDTSSSLH--------HHDDDGGDFGDDADLD----DFSGYEDAYKQ---P 822
+E+L+FL ++ H HD DF + DL+ +F + Y++ P
Sbjct: 35 DEELAFLAAEESKEQSHGGGSYHEEEHDHQHRDFENYDDLEQGGGEFHHGDHGYEEEPLP 94
Query: 821 EIDDKDVVVFKDKNFSDVVKNNRYVMVEFYAPWCGHCQALAPEYAAAATELKGENVILAK 642
+D+KDV V NF++ V NN + MVEFYAPWCG CQAL PEYAAAATELKG LAK
Sbjct: 95 PVDEKDVAVLTKDNFTEFVGNNSFAMVEFYAPWCGACQALTPEYAAAATELKGL-AALAK 153
Query: 641 VDATEESEXAQEYDIQGFPTLYFFVDG-IHKPYNGQRNKEAILTWIKKKTGPGIHNITSL 465
+DATEE + AQ+Y+IQGFPT++ FVDG + K Y G+R K+ I+TW+KKK P IHNIT+
Sbjct: 154 IDATEEGDLAQKYEIQGFPTVFLFVDGEMRKTYEGERTKDGIVTWLKKKASPSIHNITTK 213
Query: 464 EDAERILTSESKLVVGFINSLVGPESEELAAASRLEDDVNFYPTLNPDVAKLFHIDPNVK 285
E+AER+L++E KLV GF+NSLVG ESEELAAASRLEDD++FY T +PD+AKLF I+ VK
Sbjct: 214 EEAERVLSAEPKLVFGFLNSLVGSESEELAAASRLEDDLSFYQTASPDIAKLFEIETQVK 273
Query: 284 RPALILIQKEEEKLNHSGGQFRK 216
RPAL+L++KEEEKL G F K
Sbjct: 274 RPALVLLKKEEEKLARFDGNFTK 296
Score = 71.2 bits (173), Expect = 2e-11
Identities = 37/90 (41%), Positives = 54/90 (59%), Gaps = 2/90 (2%)
Frame = -1
Query: 824 PEIDDKDVVVFKDKNFSDVVKN-NRYVMVEFYAPWCGHCQALAPEYAAAATELKG-ENVI 651
PE +D DV V NF ++V + ++ V++E YAPWCGHCQ+ P Y LKG ++++
Sbjct: 432 PENNDGDVKVIVGNNFDEIVLDESKDVLLEIYAPWCGHCQSFEPIYNKLGKYLKGIDSLV 491
Query: 650 LAKVDATEESEXAQEYDIQGFPTLYFFVDG 561
+AK+D T + D GFPT+ FF G
Sbjct: 492 VAKMDGTSNEHPRAKAD--GFPTILFFPGG 519
Score = 38.9 bits (89), Expect(2) = 2e-78
Identities = 22/63 (34%), Positives = 34/63 (53%)
Frame = -2
Query: 226 NFVKSEIADFVSSNQLPLVTIFTRESAPAVSCKSNPETVAAVCNFN*LRQNPPILSRSSQ 47
NF K+ IA+FVS+N++PLV FTRE A + S + N ++ P L ++
Sbjct: 293 NFTKTAIAEFVSANKVPLVINFTREGASLIFESSVKNQLILFAKANESEKHLPTLREVAK 352
Query: 46 VFQ 38
F+
Sbjct: 353 SFK 355
>pir||T06724 protein disulfide-isomerase homolog F28P10.60 - Arabidopsis
thaliana gi|4678297|emb|CAB41088.1| protein
disulfide-isomerase-like protein [Arabidopsis thaliana]
Length = 566
Score = 277 bits (709), Expect(2) = 3e-78
Identities = 144/263 (54%), Positives = 185/263 (69%), Gaps = 16/263 (6%)
Frame = -1
Query: 956 EEDLSFLDEPDTSSSLH--------HHDDDGGDFGDDADLD----DFSGYEDAYKQ---P 822
+E+L+FL ++ H HD DF + DL+ +F + Y++ P
Sbjct: 35 DEELAFLAAEESKEQSHGGGSYHEEEHDHQHRDFENYDDLEQGGGEFHHGDHGYEEEPLP 94
Query: 821 EIDDKDVVVFKDKNFSDVVKNNRYVMVEFYAPWCGHCQALAPEYAAAATELKGENVILAK 642
+D+KDV V NF++ V NN + MVEFYAPWCG CQAL PEYAAAATELKG LAK
Sbjct: 95 PVDEKDVAVLTKDNFTEFVGNNSFAMVEFYAPWCGACQALTPEYAAAATELKGL-AALAK 153
Query: 641 VDATEESEXAQEYDIQGFPTLYFFVDG-IHKPYNGQRNKEAILTWIKKKTGPGIHNITSL 465
+DATEE + AQ+Y+IQGFPT++ FVDG + K Y G+R K+ I+TW+KKK P IHNIT+
Sbjct: 154 IDATEEGDLAQKYEIQGFPTVFLFVDGEMRKTYEGERTKDGIVTWLKKKASPSIHNITTK 213
Query: 464 EDAERILTSESKLVVGFINSLVGPESEELAAASRLEDDVNFYPTLNPDVAKLFHIDPNVK 285
E+AER+L++E KLV GF+NSLVG ESEELAAASRLEDD++FY T +PD+AKLF I+ VK
Sbjct: 214 EEAERVLSAEPKLVFGFLNSLVGSESEELAAASRLEDDLSFYQTASPDIAKLFEIETQVK 273
Query: 284 RPALILIQKEEEKLNHSGGQFRK 216
RPAL+L++KEEEKL G F K
Sbjct: 274 RPALVLLKKEEEKLARFDGNFTK 296
Score = 47.4 bits (111), Expect = 4e-04
Identities = 31/90 (34%), Positives = 48/90 (52%), Gaps = 2/90 (2%)
Frame = -1
Query: 824 PEIDDKDVVVFKDKNFSDVVKN-NRYVMVEFYAPWCGHCQALAPEYAAAATELKG-ENVI 651
PE +D DV V NF ++V + ++ V++E HCQ+ P Y LKG ++++
Sbjct: 426 PENNDGDVKVIVGNNFDEIVLDESKDVLLE-------HCQSFEPIYNKLGKYLKGIDSLV 478
Query: 650 LAKVDATEESEXAQEYDIQGFPTLYFFVDG 561
+AK+D T + D GFPT+ FF G
Sbjct: 479 VAKMDGTSNEHPRAKAD--GFPTILFFPGG 506
Score = 38.1 bits (87), Expect(2) = 3e-78
Identities = 17/30 (56%), Positives = 23/30 (76%)
Frame = -2
Query: 226 NFVKSEIADFVSSNQLPLVTIFTRESAPAV 137
NF K+ IA+FVS+N++PLV FTRE A +
Sbjct: 293 NFTKTAIAEFVSANKVPLVINFTREGASLI 322
>gb|AAO26314.1| protein disulphide isomerase [Elaeis guineensis]
Length = 447
Score = 233 bits (594), Expect(2) = 2e-67
Identities = 110/160 (68%), Positives = 140/160 (86%)
Frame = -1
Query: 695 EYAAAATELKGENVILAKVDATEESEXAQEYDIQGFPTLYFFVDGIHKPYNGQRNKEAIL 516
EYAAAAT L+GE+V LAKVDATEE+E AQ+Y++QGFPT+ FFVDG+HK Y GQR K+AI+
Sbjct: 1 EYAAAATALRGEDVALAKVDATEENELAQKYEVQGFPTVLFFVDGVHKDYPGQRTKDAIV 60
Query: 515 TWIKKKTGPGIHNITSLEDAERILTSESKLVVGFINSLVGPESEELAAASRLEDDVNFYP 336
TWIKKK GPGI NIT++E+AE ILT+E+K+V+GF+NSL G +S+ELAAAS+LEDDVNFY
Sbjct: 61 TWIKKKIGPGIQNITTVEEAENILTAENKVVLGFLNSLTGADSQELAAASKLEDDVNFYQ 120
Query: 335 TLNPDVAKLFHIDPNVKRPALILIQKEEEKLNHSGGQFRK 216
T++P VAKLFHI+P KRP+L+L++KE EKL++ GQF K
Sbjct: 121 TVSPAVAKLFHINPEAKRPSLVLLKKEAEKLSYFDGQFTK 160
Score = 78.2 bits (191), Expect = 2e-13
Identities = 40/90 (44%), Positives = 56/90 (61%), Gaps = 2/90 (2%)
Frame = -1
Query: 824 PEIDDKDVVVFKDKNFSDVVKN-NRYVMVEFYAPWCGHCQALAPEYAAAATELKG-ENVI 651
PE +D DV + NF ++V + ++ V++E YAPWCGHCQAL P Y A L+G E+++
Sbjct: 296 PETNDGDVKIVVGNNFDEIVLDESKDVLLEIYAPWCGHCQALEPTYNKLAKHLRGIESLV 355
Query: 650 LAKVDATEESEXAQEYDIQGFPTLYFFVDG 561
+AK+D T + D GFPTL FF G
Sbjct: 356 IAKMDGTSNEHPRAKVD--GFPTLLFFPAG 383
Score = 45.8 bits (107), Expect(2) = 2e-67
Identities = 19/37 (51%), Positives = 29/37 (78%)
Frame = -3
Query: 129 NPIQKQLLLFATSTDSDKILPFFLEAAKFFSQLILYM 19
NPI+KQ+LLFA S D++K++P F EAAK F ++++
Sbjct: 189 NPIKKQILLFAVSNDTEKVMPAFPEAAKLFKGKLIFV 225
Score = 39.3 bits (90), Expect = 0.098
Identities = 17/42 (40%), Positives = 27/42 (63%)
Frame = -2
Query: 262 RKKRKS*TILVANFVKSEIADFVSSNQLPLVTIFTRESAPAV 137
+K+ + + F K+ I DF+ +N+LPLV FTRE+AP +
Sbjct: 145 KKEAEKLSYFDGQFTKTAIVDFIFANKLPLVNTFTRETAPLI 186
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 881,142,049
Number of Sequences: 1393205
Number of extensions: 20395307
Number of successful extensions: 146971
Number of sequences better than 10.0: 2005
Number of HSP's better than 10.0 without gapping: 90198
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 129240
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 58773479151
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)