Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
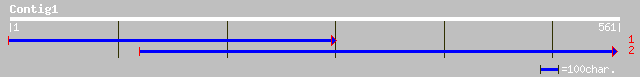
Query= KMC012856A_C01 KMC012856A_c01
(561 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P41346|FENR_VICFA Ferredoxin--NADP reductase, chloroplast pre... 298 4e-80
ref|NP_201420.1| ferredoxin-NADP+ reductase; protein id: At5g661... 297 5e-80
sp|P10933|FENR_PEA Ferredoxin--NADP reductase, leaf isozyme, chl... 296 2e-79
emb|CAB52472.1| ferredoxin-NADP+ reductase [Arabidopsis thaliana] 295 3e-79
emb|CAB71293.1| chloroplast ferredoxin-NADP+ oxidoreductase prec... 293 1e-78
>sp|P41346|FENR_VICFA Ferredoxin--NADP reductase, chloroplast precursor (FNR)
gi|551131|gb|AAA21758.1| ferredoxin NADP+ reductase
precursor
Length = 363
Score = 298 bits (762), Expect = 4e-80
Identities = 149/189 (78%), Positives = 167/189 (87%), Gaps = 2/189 (1%)
Frame = +1
Query: 1 MAAAVTAAVSFPSTKSTSLTSRTSIVAPERVVFKKVPLQYTS--GRVVSIRAQVTTDTDT 174
MAAAVTAAVS P + STSL RTSIVAPER+VFKKV L S GRV +IRAQ+TT+ +
Sbjct: 1 MAAAVTAAVSLPYSNSTSLPIRTSIVAPERLVFKKVSLNSVSISGRVGTIRAQITTEAE- 59
Query: 175 QAPPAKVEKVHKKQEEGVVVNKFKPKNPYIGRVLLNSKITGDDAPGETWHMVFSTEGEVP 354
AP KV K KKQ+EG+VVNKFKPK PY+GR LLN+KITGDDAPGETWHMVF+TEGEVP
Sbjct: 60 -APVTKVVKHSKKQDEGIVVNKFKPKEPYVGRCLLNTKITGDDAPGETWHMVFTTEGEVP 118
Query: 355 YREGQSMGVIPEGIDKNGKPHKLRLYSIASSALGDFGDSKTVSLCVKRLVYTNDSGEIVK 534
YREGQS+G++P+GIDKNGKPHKLRLYSIASSA+GDFGDSKTVSLCVKRLVYTND+GE+VK
Sbjct: 119 YREGQSIGIVPDGIDKNGKPHKLRLYSIASSAIGDFGDSKTVSLCVKRLVYTNDAGEVVK 178
Query: 535 GVCSNFLCD 561
GVCSNFLCD
Sbjct: 179 GVCSNFLCD 187
>ref|NP_201420.1| ferredoxin-NADP+ reductase; protein id: At5g66190.1, supported by
cDNA: gi_18175824, supported by cDNA: gi_20465660
[Arabidopsis thaliana] gi|10177134|dbj|BAB10424.1|
ferredoxin-NADP+ reductase [Arabidopsis thaliana]
gi|18175825|gb|AAL59934.1| putative ferredoxin-NADP+
reductase [Arabidopsis thaliana]
gi|20465661|gb|AAM20299.1| putative ferredoxin-NADP+
reductase [Arabidopsis thaliana]
Length = 360
Score = 297 bits (761), Expect = 5e-80
Identities = 148/187 (79%), Positives = 167/187 (89%)
Frame = +1
Query: 1 MAAAVTAAVSFPSTKSTSLTSRTSIVAPERVVFKKVPLQYTSGRVVSIRAQVTTDTDTQA 180
MAAA++AAVS PS+KS+SL ++ S V+P+R+ KK + Y RVVS++AQVTTDT T+A
Sbjct: 1 MAAAISAAVSLPSSKSSSLLTKISSVSPQRIFLKKSTVCYR--RVVSVKAQVTTDT-TEA 57
Query: 181 PPAKVEKVHKKQEEGVVVNKFKPKNPYIGRVLLNSKITGDDAPGETWHMVFSTEGEVPYR 360
PP KV K KKQEEG+VVNKFKPKNPY GR LLN+KITGDDAPGETWH+VF+TEGEVPYR
Sbjct: 58 PPVKVVKESKKQEEGIVVNKFKPKNPYTGRCLLNTKITGDDAPGETWHIVFTTEGEVPYR 117
Query: 361 EGQSMGVIPEGIDKNGKPHKLRLYSIASSALGDFGDSKTVSLCVKRLVYTNDSGEIVKGV 540
EGQS+GVIPEGIDKNGKPHKLRLYSIASSA+GDFGDSKTVSLCVKRLVYTND GEIVKGV
Sbjct: 118 EGQSIGVIPEGIDKNGKPHKLRLYSIASSAIGDFGDSKTVSLCVKRLVYTNDGGEIVKGV 177
Query: 541 CSNFLCD 561
CSNFLCD
Sbjct: 178 CSNFLCD 184
>sp|P10933|FENR_PEA Ferredoxin--NADP reductase, leaf isozyme, chloroplast precursor
(FNR) gi|81898|pir||S04030 ferredoxin-NADP reductase (EC
1.18.1.2) precursor - garden pea
gi|20722|emb|CAA30978.1| ferredoxin-NADH+ reductase
preprotein (AA -52 to 308) [Pisum sativum]
gi|226545|prf||1601517A ferredoxin NADP reductase
Length = 360
Score = 296 bits (757), Expect = 2e-79
Identities = 150/189 (79%), Positives = 165/189 (86%), Gaps = 2/189 (1%)
Frame = +1
Query: 1 MAAAVTAAVSFPSTKSTSLTSRTSIVAPERVVFKKVPLQYTS--GRVVSIRAQVTTDTDT 174
MAAAVTAAVS P + STSL RTSIVAPER+VFKKV L S GRV +IRAQVTT+
Sbjct: 1 MAAAVTAAVSLPYSNSTSLPIRTSIVAPERLVFKKVSLNNVSISGRVGTIRAQVTTEA-- 58
Query: 175 QAPPAKVEKVHKKQEEGVVVNKFKPKNPYIGRVLLNSKITGDDAPGETWHMVFSTEGEVP 354
PAKV K KKQ+E +VVNKFKPK PY+GR LLN+KITGDDAPGETWHMVFSTEGEVP
Sbjct: 59 ---PAKVVKHSKKQDENIVVNKFKPKEPYVGRCLLNTKITGDDAPGETWHMVFSTEGEVP 115
Query: 355 YREGQSMGVIPEGIDKNGKPHKLRLYSIASSALGDFGDSKTVSLCVKRLVYTNDSGEIVK 534
YREGQS+G++P+GIDKNGKPHKLRLYSIASSA+GDFGDSKTVSLCVKRLVYTND+GE+VK
Sbjct: 116 YREGQSIGIVPDGIDKNGKPHKLRLYSIASSAIGDFGDSKTVSLCVKRLVYTNDAGEVVK 175
Query: 535 GVCSNFLCD 561
GVCSNFLCD
Sbjct: 176 GVCSNFLCD 184
>emb|CAB52472.1| ferredoxin-NADP+ reductase [Arabidopsis thaliana]
Length = 360
Score = 295 bits (754), Expect = 3e-79
Identities = 147/187 (78%), Positives = 166/187 (88%)
Frame = +1
Query: 1 MAAAVTAAVSFPSTKSTSLTSRTSIVAPERVVFKKVPLQYTSGRVVSIRAQVTTDTDTQA 180
MAAA++AAVS PS+KS+SL ++ S V+P+R+ KK + Y RVVS++AQVTTDT T+A
Sbjct: 1 MAAAISAAVSLPSSKSSSLLTKISSVSPQRIFLKKSTVCYR--RVVSVKAQVTTDT-TEA 57
Query: 181 PPAKVEKVHKKQEEGVVVNKFKPKNPYIGRVLLNSKITGDDAPGETWHMVFSTEGEVPYR 360
PP KV K KKQEEG+VVNKFKPKNPY GR LLN+KITGDDAPGETWH+VF+TEG VPYR
Sbjct: 58 PPVKVVKESKKQEEGIVVNKFKPKNPYTGRCLLNTKITGDDAPGETWHIVFTTEGGVPYR 117
Query: 361 EGQSMGVIPEGIDKNGKPHKLRLYSIASSALGDFGDSKTVSLCVKRLVYTNDSGEIVKGV 540
EGQS+GVIPEGIDKNGKPHKLRLYSIASSA+GDFGDSKTVSLCVKRLVYTND GEIVKGV
Sbjct: 118 EGQSIGVIPEGIDKNGKPHKLRLYSIASSAIGDFGDSKTVSLCVKRLVYTNDGGEIVKGV 177
Query: 541 CSNFLCD 561
CSNFLCD
Sbjct: 178 CSNFLCD 184
>emb|CAB71293.1| chloroplast ferredoxin-NADP+ oxidoreductase precursor [Capsicum
annuum]
Length = 362
Score = 293 bits (750), Expect = 1e-78
Identities = 146/191 (76%), Positives = 165/191 (85%), Gaps = 4/191 (2%)
Frame = +1
Query: 1 MAAAVTAAVSFPSTKSTSLTSRTSIVAPERVVFKKVPLQYTS----GRVVSIRAQVTTDT 168
MA AVTAAVS PS+KSTS +RTSI++PE++ F KVPL Y + ++V+IRAQVTT+
Sbjct: 1 MATAVTAAVSLPSSKSTSFPTRTSIISPEKINFNKVPLYYRNVSGGSKLVTIRAQVTTEA 60
Query: 169 DTQAPPAKVEKVHKKQEEGVVVNKFKPKNPYIGRVLLNSKITGDDAPGETWHMVFSTEGE 348
PAKVEK+ KKQ+EGVVVNKF+PK PYIGR LLN+KITGDDAPGETWHMVFSTEGE
Sbjct: 61 -----PAKVEKISKKQDEGVVVNKFRPKEPYIGRCLLNTKITGDDAPGETWHMVFSTEGE 115
Query: 349 VPYREGQSMGVIPEGIDKNGKPHKLRLYSIASSALGDFGDSKTVSLCVKRLVYTNDSGEI 528
+PYREGQS+GVI +G+D NGKPHKLRLYSIASSALGDFGDSKTVSLCVKRLVYTND GE
Sbjct: 116 IPYREGQSIGVIADGVDANGKPHKLRLYSIASSALGDFGDSKTVSLCVKRLVYTNDKGEE 175
Query: 529 VKGVCSNFLCD 561
VKGVCSNFLCD
Sbjct: 176 VKGVCSNFLCD 186
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.314 0.131 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 539,927,828
Number of Sequences: 1393205
Number of extensions: 13268953
Number of successful extensions: 39159
Number of sequences better than 10.0: 141
Number of HSP's better than 10.0 without gapping: 36052
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38883
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)