Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
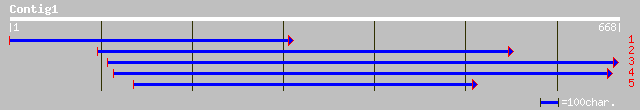
Query= KMC012850A_C01 KMC012850A_c01
(655 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG31076.1|AF283566_1 sucrose-phosphatase [Medicago truncatula] 199 2e-50
gb|AAO33160.1|AF493563_1 sucrose-phosphatase [Lycopersicon escul... 125 7e-28
pir||F84773 hypothetical protein At2g35840 [imported] - Arabidop... 122 4e-27
gb|AAG31075.1|AF283565_1 sucrose-phosphatase [Arabidopsis thaliana] 122 4e-27
ref|NP_565828.1| expressed protein; protein id: At2g35840.1, sup... 122 4e-27
>gb|AAG31076.1|AF283566_1 sucrose-phosphatase [Medicago truncatula]
Length = 419
Score = 199 bits (507), Expect = 2e-50
Identities = 90/119 (75%), Positives = 101/119 (84%)
Frame = -3
Query: 653 AEVENSEMFIAALKAVTLPSGFYIHPSGTEHNLKEYVNILRKVHGNKQGTQFRIWVDSVL 474
AEVENSE+FIAA+KA T PSG YIHPSG +HNL EY+NILRK +G KQG QFRIW+D+VL
Sbjct: 301 AEVENSELFIAAIKASTDPSGVYIHPSGADHNLNEYINILRKEYGKKQGKQFRIWLDNVL 360
Query: 473 AEQVGSDTWLVKFDKWESSGEERQGCVVTAILRKDSNWYTWMHVHQTWLEQSGHGEWII 297
A Q+ SD WLVKFDKWE EER GCVVT ILRKDS+W+TWMHVHQ+WLEQSG EWII
Sbjct: 361 ATQISSDIWLVKFDKWELHDEERHGCVVTTILRKDSDWFTWMHVHQSWLEQSGQNEWII 419
>gb|AAO33160.1|AF493563_1 sucrose-phosphatase [Lycopersicon esculentum]
Length = 425
Score = 125 bits (313), Expect = 7e-28
Identities = 57/119 (47%), Positives = 77/119 (63%), Gaps = 4/119 (3%)
Frame = -3
Query: 650 EVENSEMFIAALKAVTLPSGFYIHPSGTEHNLKEYVNILRKVHGNKQGTQFRIWVDSVLA 471
E+E+SE +++ LKAV PSG ++HPSG E +L+E V H +K G Q+R+WVD VL
Sbjct: 301 EIEHSEHYLSNLKAVCRPSGTFVHPSGVEKSLQECVTTFGTCHADKHGKQYRVWVDQVLP 360
Query: 470 EQVGSDTWLVKFDKWESSGEERQGCVVTAIL----RKDSNWYTWMHVHQTWLEQSGHGE 306
QVGSD+WLV F KWE SGE R+ C+ T +L + ++ TW HVHQTWL +
Sbjct: 361 SQVGSDSWLVSFKKWELSGENRRCCITTVLLSSKNKTVADGLTWTHVHQTWLHDDASSD 419
>pir||F84773 hypothetical protein At2g35840 [imported] - Arabidopsis thaliana
Length = 347
Score = 122 bits (306), Expect = 4e-27
Identities = 57/122 (46%), Positives = 76/122 (61%), Gaps = 4/122 (3%)
Frame = -3
Query: 650 EVENSEMFIAALKAVTLPSGFYIHPSGTEHNLKEYVNILRKVHGNKQGTQFRIWVDSVLA 471
EVENSE + A+LKA P G ++HPSGTE +L++ ++ LRK HG+KQG +FR+W D VLA
Sbjct: 226 EVENSEAYTASLKASVHPGGVFVHPSGTEKSLRDTIDELRKYHGDKQGKKFRVWADQVLA 285
Query: 470 EQVGSDTWLVKFDKWESSGEERQGCVVTA-ILRKDSNWYTWMHVHQTWLEQS---GHGEW 303
TW+VK DKWE G+ER+ C T K+ W HV QTW +++ W
Sbjct: 286 TDTTPGTWIVKLDKWEQDGDERRCCTTTVKFTSKEGEGLVWEHVQQTWSKETMVKDDSSW 345
Query: 302 II 297
II
Sbjct: 346 II 347
>gb|AAG31075.1|AF283565_1 sucrose-phosphatase [Arabidopsis thaliana]
Length = 420
Score = 122 bits (306), Expect = 4e-27
Identities = 57/122 (46%), Positives = 76/122 (61%), Gaps = 4/122 (3%)
Frame = -3
Query: 650 EVENSEMFIAALKAVTLPSGFYIHPSGTEHNLKEYVNILRKVHGNKQGTQFRIWVDSVLA 471
EVENSE + A+LKA P G ++HPSGTE +L++ ++ LRK HG+KQG +FR+W D VLA
Sbjct: 299 EVENSEAYTASLKASVHPGGVFVHPSGTEKSLRDTIDELRKYHGDKQGKKFRVWADQVLA 358
Query: 470 EQVGSDTWLVKFDKWESSGEERQGCVVTA-ILRKDSNWYTWMHVHQTWLEQS---GHGEW 303
TW+VK DKWE G+ER+ C T K+ W HV QTW +++ W
Sbjct: 359 TDTTPGTWIVKLDKWEQDGDERRCCTTTVKFTSKEGEGLVWEHVQQTWSKETMVKDDSSW 418
Query: 302 II 297
II
Sbjct: 419 II 420
>ref|NP_565828.1| expressed protein; protein id: At2g35840.1, supported by cDNA:
gi_15450787 [Arabidopsis thaliana]
gi|15450788|gb|AAK96665.1| Unknown protein [Arabidopsis
thaliana] gi|20197999|gb|AAD21473.2| expressed protein
[Arabidopsis thaliana] gi|21387099|gb|AAM47953.1|
unknown protein [Arabidopsis thaliana]
Length = 422
Score = 122 bits (306), Expect = 4e-27
Identities = 57/122 (46%), Positives = 76/122 (61%), Gaps = 4/122 (3%)
Frame = -3
Query: 650 EVENSEMFIAALKAVTLPSGFYIHPSGTEHNLKEYVNILRKVHGNKQGTQFRIWVDSVLA 471
EVENSE + A+LKA P G ++HPSGTE +L++ ++ LRK HG+KQG +FR+W D VLA
Sbjct: 301 EVENSEAYTASLKASVHPGGVFVHPSGTEKSLRDTIDELRKYHGDKQGKKFRVWADQVLA 360
Query: 470 EQVGSDTWLVKFDKWESSGEERQGCVVTA-ILRKDSNWYTWMHVHQTWLEQS---GHGEW 303
TW+VK DKWE G+ER+ C T K+ W HV QTW +++ W
Sbjct: 361 TDTTPGTWIVKLDKWEQDGDERRCCTTTVKFTSKEGEGLVWEHVQQTWSKETMVKDDSSW 420
Query: 302 II 297
II
Sbjct: 421 II 422
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 596,577,510
Number of Sequences: 1393205
Number of extensions: 13632090
Number of successful extensions: 51397
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 49105
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 51374
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 28144814643
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)