Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC012819A_C01 KMC012819A_c01
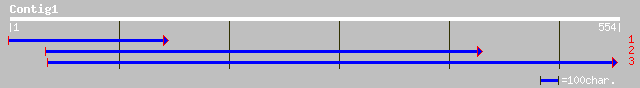
(553 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_566287.1| bHLH protein; protein id: At3g06590.1, supporte... 137 7e-32
gb|AAM67062.1| unknown [Arabidopsis thaliana] 137 9e-32
ref|NP_566567.1| bHLH protein; protein id: At3g17100.1, supporte... 134 7e-31
ref|NP_563839.1| bHLH protein; protein id: At1g09250.1, supporte... 112 2e-24
ref|NP_566260.1| bHLH protein; protein id: At3g05800.1, supporte... 93 2e-18
>ref|NP_566287.1| bHLH protein; protein id: At3g06590.1, supported by cDNA: 9391.,
supported by cDNA: gi_19698882, supported by cDNA:
gi_20260083 [Arabidopsis thaliana]
gi|12322686|gb|AAG51338.1|AC020580_18 unknown protein;
31055-31720 [Arabidopsis thaliana]
gi|19698883|gb|AAL91177.1| unknown protein [Arabidopsis
thaliana] gi|20260084|gb|AAM13389.1| unknown protein
[Arabidopsis thaliana]
Length = 221
Score = 137 bits (346), Expect = 7e-32
Identities = 85/138 (61%), Positives = 94/138 (67%), Gaps = 7/138 (5%)
Frame = -3
Query: 551 ADRALAVAARGRTRWSRAILTNRIKIKFRKQQQQQRKRLLQQKPA-------GPGRSKKA 393
ADRALAV+ARGRT WSRAIL NRIK+KFRKQ+ R R PA RS+K
Sbjct: 93 ADRALAVSARGRTLWSRAILANRIKLKFRKQR---RPRATMAIPAMTTVVSSSSNRSRKR 149
Query: 392 RFSVLRLKGKTLPAVQRKVRFLGRLVPGCRKEPLPVILEEAIDYIPALEMQVRAMAALAD 213
R SVLRL K++P V RKVR LGRLVPGC K+ +PVILEEA DYI ALEMQVRAM +L
Sbjct: 150 RVSVLRLNKKSIPDVNRKVRVLGRLVPGCGKQSVPVILEEATDYIQALEMQVRAMNSLVQ 209
Query: 212 LLLGSSSSGGGSSSFPPP 159
LL SS G S PPP
Sbjct: 210 LL---SSYG----SAPPP 220
>gb|AAM67062.1| unknown [Arabidopsis thaliana]
Length = 221
Score = 137 bits (345), Expect = 9e-32
Identities = 85/138 (61%), Positives = 94/138 (67%), Gaps = 7/138 (5%)
Frame = -3
Query: 551 ADRALAVAARGRTRWSRAILTNRIKIKFRKQQQQQRKRLLQQKPA-------GPGRSKKA 393
ADRALAV+ARGRT WSRAIL NRIK+KFRKQ+ R R PA RS+K
Sbjct: 93 ADRALAVSARGRTLWSRAILANRIKLKFRKQR---RPRATVAVPAMTTVVSSSSNRSRKR 149
Query: 392 RFSVLRLKGKTLPAVQRKVRFLGRLVPGCRKEPLPVILEEAIDYIPALEMQVRAMAALAD 213
R SVLRL K++P V RKVR LGRLVPGC K+ +PVILEEA DYI ALEMQVRAM +L
Sbjct: 150 RVSVLRLNKKSIPDVNRKVRVLGRLVPGCGKQSVPVILEEATDYIQALEMQVRAMNSLVQ 209
Query: 212 LLLGSSSSGGGSSSFPPP 159
LL SS G S PPP
Sbjct: 210 LL---SSYG----SAPPP 220
>ref|NP_566567.1| bHLH protein; protein id: At3g17100.1, supported by cDNA:
gi_15010765, supported by cDNA: gi_16323263 [Arabidopsis
thaliana] gi|7670034|dbj|BAA94988.1|
gb|AAC24081.1~gene_id:K14A17.17~similar to unknown
protein [Arabidopsis thaliana]
gi|15010766|gb|AAK74042.1| AT3g17100/K14A17_22_
[Arabidopsis thaliana] gi|16323264|gb|AAL15366.1|
AT3g17100/K14A17_22_ [Arabidopsis thaliana]
Length = 230
Score = 134 bits (337), Expect = 7e-31
Identities = 75/133 (56%), Positives = 91/133 (68%), Gaps = 1/133 (0%)
Frame = -3
Query: 551 ADRALAVAARGRTRWSRAILTNRIKIKFRKQQQQQRKR-LLQQKPAGPGRSKKARFSVLR 375
ADRALAVAARG+T WSRAIL+ +K+KFRK ++Q+ G RSKK R +VLR
Sbjct: 95 ADRALAVAARGKTLWSRAILSKAVKLKFRKHKRQRISNPTTTTLTTGSIRSKKQRATVLR 154
Query: 374 LKGKTLPAVQRKVRFLGRLVPGCRKEPLPVILEEAIDYIPALEMQVRAMAALADLLLGSS 195
LK K LPAVQRKV+ L RLVPGCRK+ LPV+LEE DYI A+EMQ+R M A+ +
Sbjct: 155 LKAKGLPAVQRKVKVLSRLVPGCRKQSLPVVLEETTDYIAAMEMQIRTMTAILSAV---- 210
Query: 194 SSGGGSSSFPPPS 156
SSS PPP+
Sbjct: 211 -----SSSPPPPT 218
>ref|NP_563839.1| bHLH protein; protein id: At1g09250.1, supported by cDNA: 9228.
[Arabidopsis thaliana] gi|25402527|pir||D86225
hypothetical protein [imported] - Arabidopsis thaliana
gi|3249098|gb|AAC24081.1| ESTs gb|T04610, gb|N38459,
gb|T45174, gb|R30481 and gb|N64971 come from this gene.
[Arabidopsis thaliana] gi|27764992|gb|AAO23617.1|
At1g09250 [Arabidopsis thaliana]
Length = 207
Score = 112 bits (281), Expect = 2e-24
Identities = 62/119 (52%), Positives = 82/119 (68%)
Frame = -3
Query: 551 ADRALAVAARGRTRWSRAILTNRIKIKFRKQQQQQRKRLLQQKPAGPGRSKKARFSVLRL 372
ADR LA +ARG TRWSRAIL +R++ K +K ++ + K G +S+K R+
Sbjct: 91 ADRVLAASARGTTRWSRAILASRVRAKLKKHRKAK-------KSTGNCKSRKGLTETNRI 143
Query: 371 KGKTLPAVQRKVRFLGRLVPGCRKEPLPVILEEAIDYIPALEMQVRAMAALADLLLGSS 195
K LPAV+RK++ LGRLVPGCRK +P +L+EA DYI ALEMQVRAM ALA+LL ++
Sbjct: 144 K---LPAVERKLKILGRLVPGCRKVSVPNLLDEATDYIAALEMQVRAMEALAELLTAAA 199
>ref|NP_566260.1| bHLH protein; protein id: At3g05800.1, supported by cDNA: 21672.
[Arabidopsis thaliana]
gi|6714393|gb|AAF26082.1|AC012393_8 hypothetical protein
[Arabidopsis thaliana] gi|21554259|gb|AAM63334.1|
unknown [Arabidopsis thaliana]
Length = 211
Score = 93.2 bits (230), Expect = 2e-18
Identities = 54/115 (46%), Positives = 67/115 (57%)
Frame = -3
Query: 551 ADRALAVAARGRTRWSRAILTNRIKIKFRKQQQQQRKRLLQQKPAGPGRSKKARFSVLRL 372
AD+ LA ARG TRWSRAIL +R R+++ + L G G S + R
Sbjct: 93 ADKVLATTARGATRWSRAILVSRFGTSLRRRRNTKPASALAAAIRGSGGSGRRR------ 146
Query: 371 KGKTLPAVQRKVRFLGRLVPGCRKEPLPVILEEAIDYIPALEMQVRAMAALADLL 207
L AV +VR LG LVPGCR+ LP +L+E DYI ALEMQVRAM AL+ +L
Sbjct: 147 ---KLSAVGNRVRVLGGLVPGCRRTALPELLDETADYIAALEMQVRAMTALSKIL 198
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 546,628,324
Number of Sequences: 1393205
Number of extensions: 14080595
Number of successful extensions: 113898
Number of sequences better than 10.0: 667
Number of HSP's better than 10.0 without gapping: 73228
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 105036
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19234190289
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)