Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
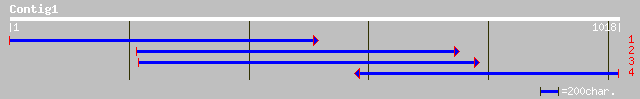
Query= KMC012809A_C01 KMC012809A_c01
(1018 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO43000.1| early tobacco anther 1 [Nicotiana tabacum] 165 9e-40
ref|NP_568698.1| expressed protein; protein id: At5g48480.1, sup... 107 3e-22
ref|NP_436346.1| Hypothetical protein [Sinorhizobium meliloti] g... 56 8e-07
ref|NP_745582.1| glyoxalase family protein [Pseudomonas putida K... 55 1e-06
ref|NP_518228.1| CONSERVED HYPOTHETICAL PROTEIN [Ralstonia solan... 55 2e-06
>gb|AAO43000.1| early tobacco anther 1 [Nicotiana tabacum]
Length = 166
Score = 165 bits (418), Expect = 9e-40
Identities = 87/131 (66%), Positives = 99/131 (75%), Gaps = 1/131 (0%)
Frame = +3
Query: 366 EAPKANDAVLFYKAAFGAEEVSRALNPKRKADHELPLILSAELKIAGSSILVSDLAVD-S 542
EAPKA DA FYKAAFG EEV+R +N KRKA+ E+PL+ S ELK+ SS LVS+L D S
Sbjct: 36 EAPKAKDAAQFYKAAFGVEEVTR-VNQKRKAEQEIPLVSSVELKLGSSSFLVSNLTDDDS 94
Query: 543 TAPAKTGGNGIVLCLETEDVEAAVAKAVRAGAVKEGEVVEGEGGACCGGLVVKVKDPYGF 722
+AP K G V CLETEDVEAAVAKAV AGAV EGE+ EG+G A GG V K+KDPYG+
Sbjct: 95 SAPVKAATTGCVFCLETEDVEAAVAKAVSAGAVSEGEIAEGDGAAYFGGRVGKLKDPYGY 154
Query: 723 IWQICSPVKAS 755
IW ICSPVK S
Sbjct: 155 IWMICSPVKKS 165
>ref|NP_568698.1| expressed protein; protein id: At5g48480.1, supported by cDNA:
31667., supported by cDNA: gi_18389223 [Arabidopsis
thaliana] gi|8777378|dbj|BAA96968.1|
gene_id:MJE7.12~unknown protein [Arabidopsis thaliana]
gi|18389224|gb|AAL67055.1| unknown protein [Arabidopsis
thaliana] gi|21592710|gb|AAM64659.1| unknown
[Arabidopsis thaliana] gi|22136970|gb|AAM91714.1|
unknown protein [Arabidopsis thaliana]
Length = 166
Score = 107 bits (267), Expect = 3e-22
Identities = 65/125 (52%), Positives = 81/125 (64%), Gaps = 1/125 (0%)
Frame = +3
Query: 357 VLSEAPKANDAVLFYKAAFGAEEVSRALNPKRKADHELPLILSAELKIAGSSILVSDL-A 533
+L EA K DAV FYK+AFGA E +L PKRK D ELP +LS+EL +AGSS +V D+ +
Sbjct: 30 LLVEAQKVGDAVTFYKSAFGAIESGHSLYPKRKLDQELPHVLSSELNLAGSSFVVCDVSS 89
Query: 534 VDSTAPAKTGGNGIVLCLETEDVEAAVAKAVRAGAVKEGEVVEGEGGACCGGLVVKVKDP 713
+ + AK+ G+G+ L T+D EAAVAKAV AGAVK EV E E G KV DP
Sbjct: 90 LPGFSTAKSEGSGVTFLLGTKDAEAAVAKAVDAGAVKV-EVTEAEVEL---GFKGKVTDP 145
Query: 714 YGFIW 728
+G W
Sbjct: 146 FGVTW 150
>ref|NP_436346.1| Hypothetical protein [Sinorhizobium meliloti]
gi|25505078|pir||D95399 protein [imported] -
Sinorhizobium meliloti (strain 1021) magaplasmid pSymA
gi|14524256|gb|AAK65758.1| Hypothetical protein
[Sinorhizobium meliloti]
Length = 149
Score = 56.2 bits (134), Expect = 8e-07
Identities = 42/126 (33%), Positives = 63/126 (49%), Gaps = 2/126 (1%)
Frame = +3
Query: 378 ANDAVLFYKAAFGAEEVSRALNPKRKADHELPLILSAELKIAGSSILVSDLAVD--STAP 551
A A+ FY AFGAEE+ R +P+ I AELKI S+++++D D + +P
Sbjct: 17 ARVAIDFYGRAFGAEELFRMTDPRDGR------IGHAELKIGESTVMIADEYPDFGALSP 70
Query: 552 AKTGGNGIVLCLETEDVEAAVAKAVRAGAVKEGEVVEGEGGACCGGLVVKVKDPYGFIWQ 731
GG+ + L T V+A +A+AV AGAV + G V V DP+G W
Sbjct: 71 DTIGGSPVTFHLATLAVDADLARAVDAGAV----ALRAAADQGYGERVAMVVDPFGHRWM 126
Query: 732 ICSPVK 749
+ ++
Sbjct: 127 LSQKIE 132
>ref|NP_745582.1| glyoxalase family protein [Pseudomonas putida KT2440]
gi|24985094|gb|AAN69046.1|AE016537_4 glyoxalase family
protein [Pseudomonas putida KT2440]
Length = 159
Score = 55.5 bits (132), Expect = 1e-06
Identities = 42/120 (35%), Positives = 59/120 (49%), Gaps = 3/120 (2%)
Frame = +3
Query: 378 ANDAVLFYKAAFGAEEVSRALNPKRKADHELPLILSAELKIAGSSILVS---DLAVDSTA 548
A A+ FYK AFGA E+ R P + H AEL+I SS+++ D+ TA
Sbjct: 22 AAKAIEFYKQAFGAVEMFRLDAPGGRVGH-------AELRIGDSSLMLGDPCDMEGGLTA 74
Query: 549 PAKTGGNGIVLCLETEDVEAAVAKAVRAGAVKEGEVVEGEGGACCGGLVVKVKDPYGFIW 728
K G G+ L L ED + A+A+ AG ++ + + G G L KDP+G IW
Sbjct: 75 SQKLNGAGVGLHLYVEDCDKVYAQALAAGGIQLHPLTDQFYGDRSGTL----KDPFGNIW 130
>ref|NP_518228.1| CONSERVED HYPOTHETICAL PROTEIN [Ralstonia solanacearum]
gi|17427115|emb|CAD13635.1| CONSERVED HYPOTHETICAL
PROTEIN [Ralstonia solanacearum]
Length = 159
Score = 54.7 bits (130), Expect = 2e-06
Identities = 38/126 (30%), Positives = 60/126 (47%), Gaps = 2/126 (1%)
Frame = +3
Query: 378 ANDAVLFYKAAFGAEEVSRALNPKRKADHELPLILSAELKIAGSSILVSDLA--VDSTAP 551
A+ A+ FYK AFGA E+ R P H AE++I S ++++D + +P
Sbjct: 22 ADKALDFYKQAFGATEIMRMAGPNGAIAH-------AEIRIGDSPVMMADEMPNMSCASP 74
Query: 552 AKTGGNGIVLCLETEDVEAAVAKAVRAGAVKEGEVVEGEGGACCGGLVVKVKDPYGFIWQ 731
+ L + +DV+A A A++ GA + V G G L KDP+G +W
Sbjct: 75 DTLQSTSVGLMIYVKDVDAVFANAIKTGATEVRPVQNQFYGDRSGTL----KDPFGHVWT 130
Query: 732 ICSPVK 749
I + V+
Sbjct: 131 ISTHVE 136
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 853,150,757
Number of Sequences: 1393205
Number of extensions: 19511170
Number of successful extensions: 81285
Number of sequences better than 10.0: 262
Number of HSP's better than 10.0 without gapping: 64699
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 78583
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 59049410978
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)