Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC012806A_C01 KMC012806A_c01
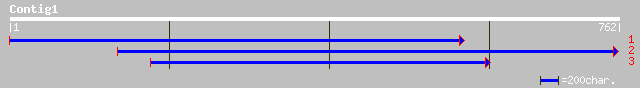
(759 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC43363.1| unknown protein [Arabidopsis thaliana] 92 8e-18
ref|NP_012331.1| ReCYcling 1; Rcy1p [Saccharomyces cerevisiae] g... 32 7.6
sp|P39531|YJU4_YEAST Hypothetical 75.9 kDa protein in NCE1-PRP21... 32 7.6
gb|AAH47805.1| Similar to kinesin family member 23 [Danio rerio] 32 7.6
ref|NP_571430.1| kinesin-like 5; mitotic kinesin-like protein 1 ... 32 7.6
>dbj|BAC43363.1| unknown protein [Arabidopsis thaliana]
Length = 258
Score = 92.0 bits (227), Expect = 8e-18
Identities = 45/72 (62%), Positives = 55/72 (75%)
Frame = -2
Query: 755 ASLYSARAIKVKEVDGFRNYYFYEFGNDQLHIALMAGVSGGKAIIAGATASQSKWDIDGV 576
A++YSAR IKVKE +G RNYYFYEFG D+ IAL+A V+ GK IAGA A +SKW D +
Sbjct: 187 ATIYSARTIKVKEEEGLRNYYFYEFGRDEERIALVASVNRGKVYIAGAAAPESKWKDDEL 246
Query: 575 RLRSAAVSLKIL 540
+LRSAA+S IL
Sbjct: 247 KLRSAAISFTIL 258
>ref|NP_012331.1| ReCYcling 1; Rcy1p [Saccharomyces cerevisiae]
gi|7493837|pir||S77615 hypothetical protein YJL204c -
yeast (Saccharomyces cerevisiae)
gi|1183993|emb|CAA54753.1| hypothetical protein J0318
[Saccharomyces cerevisiae]
Length = 840
Score = 32.3 bits (72), Expect = 7.6
Identities = 29/87 (33%), Positives = 39/87 (44%), Gaps = 1/87 (1%)
Frame = +1
Query: 16 HSASCSWQNLEGLSSFTVIMV*FQLKKNELFDQTNQTPFNLTR*KPPTST-KEVIKEEGK 192
HS S L+ LSSFT I F +N+ Q Q +NL K +I
Sbjct: 442 HSTDISNDKLDFLSSFTKIFK-FSNNENQRLKQQLQLAYNLNLISNNLQNIKSLI----- 495
Query: 193 SSDLCYRICNETQPE*TSLLKTNT*EN 273
S DLCY+I ET + + K +T E+
Sbjct: 496 SLDLCYKILQETSEKTDQIYKFHTIES 522
>sp|P39531|YJU4_YEAST Hypothetical 75.9 kDa protein in NCE1-PRP21 intergenic region
gi|2131088|emb|CAA89499.1| ORF YJL204c [Saccharomyces
cerevisiae]
Length = 653
Score = 32.3 bits (72), Expect = 7.6
Identities = 29/87 (33%), Positives = 39/87 (44%), Gaps = 1/87 (1%)
Frame = +1
Query: 16 HSASCSWQNLEGLSSFTVIMV*FQLKKNELFDQTNQTPFNLTR*KPPTST-KEVIKEEGK 192
HS S L+ LSSFT I F +N+ Q Q +NL K +I
Sbjct: 442 HSTDISNDKLDFLSSFTKIFK-FSNNENQRLKQQLQLAYNLNLISNNLQNIKSLI----- 495
Query: 193 SSDLCYRICNETQPE*TSLLKTNT*EN 273
S DLCY+I ET + + K +T E+
Sbjct: 496 SLDLCYKILQETSEKTDQIYKFHTIES 522
>gb|AAH47805.1| Similar to kinesin family member 23 [Danio rerio]
Length = 867
Score = 32.3 bits (72), Expect = 7.6
Identities = 18/73 (24%), Positives = 37/73 (50%)
Frame = +1
Query: 13 SHSASCSWQNLEGLSSFTVIMV*FQLKKNELFDQTNQTPFNLTR*KPPTSTKEVIKEEGK 192
S +C + ++ SS++V + ++ N ++D +TPF+ + KPP S I E +
Sbjct: 200 SPEEACKAEGVDEDSSYSVFVSYIEIYNNYIYDLLEETPFDPIKPKPPQSK---ILREDQ 256
Query: 193 SSDLCYRICNETQ 231
+ ++ C E +
Sbjct: 257 NHNMYVAGCTEVE 269
>ref|NP_571430.1| kinesin-like 5; mitotic kinesin-like protein 1 [Danio rerio]
gi|6006743|gb|AAF00594.1|AF139990_1 mitotic kinesin-like
protein 1 [Danio rerio]
Length = 867
Score = 32.3 bits (72), Expect = 7.6
Identities = 18/73 (24%), Positives = 37/73 (50%)
Frame = +1
Query: 13 SHSASCSWQNLEGLSSFTVIMV*FQLKKNELFDQTNQTPFNLTR*KPPTSTKEVIKEEGK 192
S +C + ++ SS++V + ++ N ++D +TPF+ + KPP S I E +
Sbjct: 200 SPEEACKAEGVDEDSSYSVFVSYIEIYNNYIYDLLEETPFDPIKPKPPQSK---ILREDQ 256
Query: 193 SSDLCYRICNETQ 231
+ ++ C E +
Sbjct: 257 NHNMYVAGCTEVE 269
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 607,982,621
Number of Sequences: 1393205
Number of extensions: 11948468
Number of successful extensions: 24356
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 23721
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24345
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 37158613404
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)