Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC012764A_C01 KMC012764A_c01
(523 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
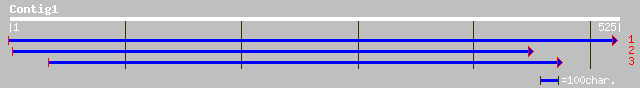
ref|NP_172035.1| hypothetical protein; protein id: At1g05440.1 [... 107 8e-23
gb|AAK71563.1|AC087852_23 hypothetical protein [Oryza sativa (ja... 79 3e-14
gb|EAA31110.1| hypothetical protein [Neurospora crassa] 36 0.31
ref|NP_040988.1| replicase [Foxtail mosaic virus] gi|139133|sp|P... 35 0.40
gb|AAN46894.1| RNA-dependent RNA polymerase [Foxtail mosaic virus] 35 0.40
>ref|NP_172035.1| hypothetical protein; protein id: At1g05440.1 [Arabidopsis
thaliana] gi|25406842|pir||B86189 protein T25N20.9
[imported] - Arabidopsis thaliana
gi|8778729|gb|AAF79737.1|AC005106_18 T25N20.9
[Arabidopsis thaliana]
Length = 393
Score = 107 bits (267), Expect = 8e-23
Identities = 50/107 (46%), Positives = 73/107 (67%), Gaps = 2/107 (1%)
Frame = -2
Query: 522 GWPLQTAGLLTNGFSTRTQTILSGRVTEWNGGLVGYLTRKANGSWVQPKWGASVVQLDPN 343
GWPLQT+GL GFS+ + T+LSGRVTEW G GY + N SW + KW SV+QLDPN
Sbjct: 269 GWPLQTSGLFHTGFSSSSITVLSGRVTEWTEGKFGYSIHETNTSWGKTKWSTSVLQLDPN 328
Query: 342 TWVLEYQRSSIV-DGTRLYSAVLEFLKYRNSRVIGRL-KKKFWQFAA 208
TWVLEY +SS++ D + L S ++ +++ RV + ++++W F++
Sbjct: 329 TWVLEYSQSSVMDDSSSLLSLTIDIMEHLVFRVAKNVNRERYWMFSS 375
>gb|AAK71563.1|AC087852_23 hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 321
Score = 79.0 bits (193), Expect = 3e-14
Identities = 43/100 (43%), Positives = 59/100 (59%), Gaps = 1/100 (1%)
Frame = -2
Query: 522 GWPLQTAGLLTNGFSTRTQTILSGRVTEWNGGLVGYLTRKANG-SWVQPKWGASVVQLDP 346
G PL+T+GLL R TILSG++TEW+ G V R +NG SW + V+L+
Sbjct: 204 GSPLRTSGLLPRALPARHLTILSGKITEWSDGRVWPTVRASNGSSWSYGGRSSPAVRLEA 263
Query: 345 NTWVLEYQRSSIVDGTRLYSAVLEFLKYRNSRVIGRLKKK 226
TWV+EYQRS + +GTRL A E + R S V R +++
Sbjct: 264 ETWVVEYQRSVVFEGTRLIPAAAELVASRCSAVARRARQR 303
>gb|EAA31110.1| hypothetical protein [Neurospora crassa]
Length = 371
Score = 35.8 bits (81), Expect = 0.31
Identities = 18/56 (32%), Positives = 30/56 (53%)
Frame = -2
Query: 459 LSGRVTEWNGGLVGYLTRKANGSWVQPKWGASVVQLDPNTWVLEYQRSSIVDGTRL 292
++G++ G V LT K +GSWV+P+ + P+ W L+ Q S++ G L
Sbjct: 1 MTGKIPTVEGIPVLPLTHKDDGSWVRPETANDAINPAPSRWYLKAQASALRSGLYL 56
>ref|NP_040988.1| replicase [Foxtail mosaic virus] gi|139133|sp|P22168|RRPO_FXMV RNA
replication protein (152 kDa protein) (ORF 1) [Contains:
RNA-directed RNA polymerase ; Probable helicase]
gi|94402|pir||JQ1258 RNA-directed RNA polymerase (EC
2.7.7.48) - foxtail mosaic virus
gi|325392|gb|AAA43826.1| 152K protein
Length = 1335
Score = 35.4 bits (80), Expect = 0.40
Identities = 22/63 (34%), Positives = 31/63 (48%)
Frame = +2
Query: 200 PSNAANCQNFFFSLPITLEFLYLRNSNTAEYNLVPSTIELLWYSRTHVLGSNCTTEAPHF 379
P N AN + P ++ LY RN T +NLVPS +++ Y+ LG +T A
Sbjct: 736 PRNLANALGVYSETPGEVKVLYTRNIKTGYHNLVPSQMKMRNYAS---LGQRASTYAGCQ 792
Query: 380 GCT 388
G T
Sbjct: 793 GIT 795
>gb|AAN46894.1| RNA-dependent RNA polymerase [Foxtail mosaic virus]
Length = 1335
Score = 35.4 bits (80), Expect = 0.40
Identities = 22/63 (34%), Positives = 31/63 (48%)
Frame = +2
Query: 200 PSNAANCQNFFFSLPITLEFLYLRNSNTAEYNLVPSTIELLWYSRTHVLGSNCTTEAPHF 379
P N AN + P ++ LY RN T +NLVPS +++ Y+ LG +T A
Sbjct: 736 PRNLANALGVYSETPGEVKVLYTRNIRTGYHNLVPSQMKMRNYAS---LGERASTYAGCQ 792
Query: 380 GCT 388
G T
Sbjct: 793 GIT 795
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 455,407,021
Number of Sequences: 1393205
Number of extensions: 9921466
Number of successful extensions: 24742
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 24098
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24736
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16731298976
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)