Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC012582A_C01 KMC012582A_c01
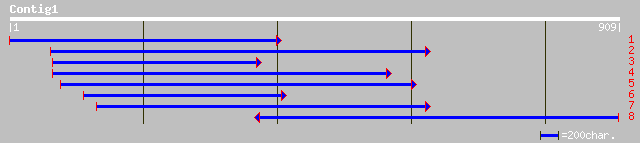
(908 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF02148.1|AC009853_8 unknown protein [Arabidopsis thaliana] 143 4e-33
gb|AAM65781.1| unknown [Arabidopsis thaliana] 137 2e-31
ref|NP_193034.1| putative protein; protein id: At4g12980.1, supp... 137 2e-31
pir||T51337 auxin-induced protein AIR12 [imported] - Arabidopsis... 134 2e-30
ref|NP_566763.1| expressed protein; protein id: At3g25290.1, sup... 126 4e-28
>gb|AAF02148.1|AC009853_8 unknown protein [Arabidopsis thaliana]
Length = 252
Score = 143 bits (360), Expect = 4e-33
Identities = 90/233 (38%), Positives = 128/233 (54%), Gaps = 12/233 (5%)
Frame = -3
Query: 852 LSLLTHSFRHQRA-TQKLPSGRNYASCTNLSSLGATLHFTYNETNSSIAVAFSAAPPK-- 682
+SL++ + Q +Q L S + SC +L L + LH+TYN +NSS++VAF A P +
Sbjct: 17 VSLISPAISQQACKSQNLNSAGPFDSCEDLPVLNSYLHYTYNSSNSSLSVAFVATPSQAN 76
Query: 681 GGWVAWGLNLAGGGMAGTQALVAYKGGDKNVVGVHLYNITSYSELAEVKSFSFETWDLSA 502
GGWVAW +N G MAG+QA +AY+ G V YNI+SYS L E K +F+ W+L A
Sbjct: 77 GGWVAWAINPTGTKMAGSQAFLAYRSGGGAAPVVKTYNISSYSSLVEGK-LAFDFWNLRA 135
Query: 501 EE-SSAAITIFAAVKVPENADNISQIWQV-GPVVSGKINKHDFKPENLAAKAPLS----V 340
E S I IF VKVP AD+++Q+WQ+ G V +G+ H F P+NL + LS
Sbjct: 136 ESLSGGRIAIFTTVKVPAGADSVNQVWQIGGNVTNGRPGVHPFGPDNLGSHRVLSFTEDA 195
Query: 339 VATTAVVGGGNTTSGSGNTTTSGQKGGGVHVMG---ERFGLGFYLGVVLVIMS 190
+A G G+ +TT G GG G G LG+++++ S
Sbjct: 196 APGSAPSPGSAPAPGTSGSTTPGTAAGGPGNAGSLTRNVNFGVNLGILVLLGS 248
>gb|AAM65781.1| unknown [Arabidopsis thaliana]
Length = 394
Score = 137 bits (346), Expect = 2e-31
Identities = 87/216 (40%), Positives = 124/216 (57%), Gaps = 11/216 (5%)
Frame = -3
Query: 816 ATQKLPSGRNYASCTNLSSLGATLHFTYNETNSSIAVAFSAAPPK-GGWVAWGLNLAGGG 640
++Q ++Y C +L L A LH++Y+ +N+++AV FSA P K GGW+AW +N G
Sbjct: 30 SSQTFSGVKSYPHCLDLPDLKAILHYSYDASNTTLAVVFSAPPSKPGGWIAWAINPKSTG 89
Query: 639 MAGTQALVAYKGGDKNVVGVHLYNITSYSELAEVKSFSFETWDLSAEESS---AAITIFA 469
MAG+QALVA K V V NI SYS L K SF+ WD+ AEE++ A+ IFA
Sbjct: 90 MAGSQALVASKDPSTGVASVTTLNIVSYSSLVPSK-LSFDVWDVKAEEAANDGGALRIFA 148
Query: 468 AVKVPEN---ADNISQIWQVGPVVS-GKINKHDFKPENLAAKAPLSVVATT---AVVGGG 310
VKVP + + ++Q+WQVGP VS G+I HDF NL + L + TT V GGG
Sbjct: 149 KVKVPADLAASGKVNQVWQVGPGVSNGRIQAHDFSGPNLNSVGSLDLTGTTPGVPVSGGG 208
Query: 309 NTTSGSGNTTTSGQKGGGVHVMGERFGLGFYLGVVL 202
G+GN+ + G+ + +GL F +G ++
Sbjct: 209 ----GAGNSRIHKRNIHGI-LNAVSWGLLFPIGAMI 239
>ref|NP_193034.1| putative protein; protein id: At4g12980.1, supported by cDNA: 4642.
[Arabidopsis thaliana] gi|7486133|pir||T10200
hypothetical protein F25G13.70 - Arabidopsis thaliana
gi|5123939|emb|CAB45497.1| putative protein [Arabidopsis
thaliana] gi|7268000|emb|CAB78340.1| putative protein
[Arabidopsis thaliana] gi|28392885|gb|AAO41879.1|
unknown protein [Arabidopsis thaliana]
gi|28827634|gb|AAO50661.1| unknown protein [Arabidopsis
thaliana]
Length = 394
Score = 137 bits (346), Expect = 2e-31
Identities = 87/216 (40%), Positives = 124/216 (57%), Gaps = 11/216 (5%)
Frame = -3
Query: 816 ATQKLPSGRNYASCTNLSSLGATLHFTYNETNSSIAVAFSAAPPK-GGWVAWGLNLAGGG 640
++Q ++Y C +L L A LH++Y+ +N+++AV FSA P K GGW+AW +N G
Sbjct: 30 SSQTFSGVKSYPHCLDLPDLKAILHYSYDASNTTLAVVFSAPPSKPGGWIAWAINPKSTG 89
Query: 639 MAGTQALVAYKGGDKNVVGVHLYNITSYSELAEVKSFSFETWDLSAEESS---AAITIFA 469
MAG+QALVA K V V NI SYS L K SF+ WD+ AEE++ A+ IFA
Sbjct: 90 MAGSQALVASKDPSTGVASVTTLNIVSYSSLVPSK-LSFDVWDVKAEEAANDGGALRIFA 148
Query: 468 AVKVPEN---ADNISQIWQVGPVVS-GKINKHDFKPENLAAKAPLSVVATT---AVVGGG 310
VKVP + + ++Q+WQVGP VS G+I HDF NL + L + TT V GGG
Sbjct: 149 KVKVPADLAASGKVNQVWQVGPGVSNGRIQAHDFSGPNLNSVGSLDLTGTTPGVPVSGGG 208
Query: 309 NTTSGSGNTTTSGQKGGGVHVMGERFGLGFYLGVVL 202
G+GN+ + G+ + +GL F +G ++
Sbjct: 209 ----GAGNSRIHKRNIHGI-LNAVSWGLLFPIGAMI 239
>pir||T51337 auxin-induced protein AIR12 [imported] - Arabidopsis thaliana
(fragment) gi|3695023|gb|AAC62613.1| unknown
[Arabidopsis thaliana]
Length = 264
Score = 134 bits (336), Expect = 2e-30
Identities = 89/240 (37%), Positives = 127/240 (52%), Gaps = 19/240 (7%)
Frame = -3
Query: 852 LSLLTHSFRHQRA-TQKLPSGRNYASCTNLSSLGATLHFTYNETNSSIAVAFSAAPPK-- 682
+SL++ + Q +Q L S + SC +L L + LH+TYN +NSS++VAF A P +
Sbjct: 22 VSLISPAISQQACKSQNLNSAGPFDSCEDLPVLNSYLHYTYNSSNSSLSVAFVATPSQAN 81
Query: 681 GGWVAWGLNLAGGGMAGTQALVAYKGGDKNVVGVHLYNITSYSELAEVKSFSFETWDLSA 502
GGWVAW +N G MAG+QA +AY+ G V YNI+SYS L E K +F+ W+L A
Sbjct: 82 GGWVAWAINPTGTKMAGSQAFLAYRSGGGAAPVVKTYNISSYSSLVEGK-LAFDFWNLRA 140
Query: 501 EE-SSAAITIF-AAVKVPENADNISQIWQV-GPVVSGKINKHDFKPENLAAKAPLSVVAT 331
E S I IF VKVP D+++Q+WQ+ G V +G+ H F P+NL + LS
Sbjct: 141 ESLSGGRIAIFNRTVKVPAGRDSVNQVWQIGGNVTNGRPGVHPFGPDNLGSHRVLSFTED 200
Query: 330 ----------TAVVGGGNTTSGSGNTTTSGQKGGGVHVMG---ERFGLGFYLGVVLVIMS 190
+A G G+ +TT G GG G G LG+++++ S
Sbjct: 201 AAPGSAPSPGSAPSPGSAPAPGTSGSTTPGTAAGGPGNAGSLTRNVNFGVNLGILVLLGS 260
>ref|NP_566763.1| expressed protein; protein id: At3g25290.1, supported by cDNA:
gi_16323199 [Arabidopsis thaliana]
gi|9294186|dbj|BAB02088.1|
emb|CAB45497.1~gene_id:MJL12.25~strong similarity to
unknown protein [Arabidopsis thaliana]
gi|16323200|gb|AAL15334.1| AT3g25290/MJL12_25
[Arabidopsis thaliana] gi|25090093|gb|AAN72226.1|
At3g25290/MJL12_25 [Arabidopsis thaliana]
Length = 393
Score = 126 bits (317), Expect = 4e-28
Identities = 78/215 (36%), Positives = 118/215 (54%), Gaps = 11/215 (5%)
Frame = -3
Query: 813 TQKLPSGRNYASCTNLSSLGATLHFTYNETNSSIAVAFSAAPPK-GGWVAWGLNLAGGGM 637
+Q + Y C +L L A LH++Y+ +N+++AV FSA P K GGW+AW +N GM
Sbjct: 30 SQTFSGDKTYPHCLDLPQLKAFLHYSYDASNTTLAVVFSAPPAKPGGWIAWAINPKATGM 89
Query: 636 AGTQALVAYKGGDKNVVGVHLYNITSYSELAEVKSFSFETWDLSAEESS---AAITIFAA 466
G+Q LVAYK V V NI+SYS L K +F+ WD+ AEE++ ++ IFA
Sbjct: 90 VGSQTLVAYKDPGNGVAVVKTLNISSYSSLIPSK-LAFDVWDMKAEEAARDGGSLRIFAR 148
Query: 465 VKVPEN---ADNISQIWQVGPVV--SGKINKHDFKPENLAAKAPLSVVATTAVVGGGNTT 301
VKVP + ++Q+WQVGP + G I +H F NLA+ + L + + G T
Sbjct: 149 VKVPADLVAKGKVNQVWQVGPELGPGGMIGRHAFDSANLASMSSLDLKGDNS----GGTI 204
Query: 300 SGSGNTTTSGQKGGGVH--VMGERFGLGFYLGVVL 202
SG G+ + K +H + +G+ F +G ++
Sbjct: 205 SG-GDEVNAKIKNRNIHGILNAVSWGILFPIGAII 238
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 930,951,049
Number of Sequences: 1393205
Number of extensions: 27326753
Number of successful extensions: 262602
Number of sequences better than 10.0: 5315
Number of HSP's better than 10.0 without gapping: 132149
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 216740
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 49641180728
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)