Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC012349A_C02 KMC012349A_c02
(611 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
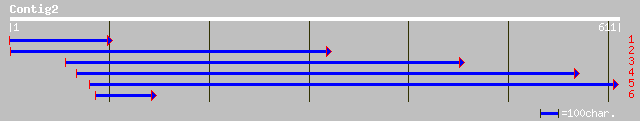
dbj|BAA81687.1| expressed in cucumber hypocotyls [Cucumis sativus] 147 8e-35
ref|NP_187124.1| auxin-induced protein; protein id: At3g04730.1,... 145 4e-34
gb|AAM96891.1| auxin-responsive protein IAA1; MjAux/IAA1 [Mirabi... 144 7e-34
gb|AAN16886.1| Aux/IAA1 [Mirabilis jalapa] 144 7e-34
gb|AAD32147.1|AF123509_1 Nt-iaa4.1 deduced protein [Nicotiana ta... 144 1e-33
>dbj|BAA81687.1| expressed in cucumber hypocotyls [Cucumis sativus]
Length = 230
Score = 147 bits (372), Expect = 8e-35
Identities = 69/71 (97%), Positives = 70/71 (98%)
Frame = -1
Query: 608 MKDFMNESKLMDLLNSSDYVPTYEDKDGDWMLVGDVPWEMFVESCKRLRIMKGKEAIGLA 429
MKDFMNESKLMDLL+ SDYVPTYEDKDGDWMLVGDVPWEMFVESCKRLRIMKGKEAIGLA
Sbjct: 160 MKDFMNESKLMDLLSGSDYVPTYEDKDGDWMLVGDVPWEMFVESCKRLRIMKGKEAIGLA 219
Query: 428 PRAMEKCKNRS 396
PRAMEKCKNRS
Sbjct: 220 PRAMEKCKNRS 230
>ref|NP_187124.1| auxin-induced protein; protein id: At3g04730.1, supported by cDNA:
32727., supported by cDNA: gi_12083209, supported by
cDNA: gi_14030658, supported by cDNA: gi_2618720
[Arabidopsis thaliana] gi|11131089|sp|O24407|AXIG_ARATH
Auxin-responsive protein IAA16 (Indoleacetic
acid-induced protein 16) gi|2618721|gb|AAB84353.1| IAA16
[Arabidopsis thaliana]
gi|6175173|gb|AAF04899.1|AC011437_14 auxin-induced
protein [Arabidopsis thaliana]
gi|12083210|gb|AAG48764.1|AF332400_1 putative
auxin-induced protein IAA16 [Arabidopsis thaliana]
gi|14030659|gb|AAK53004.1|AF375420_1 AT3g04730/F7O18_22
[Arabidopsis thaliana] gi|21592802|gb|AAM64751.1|
auxin-induced protein [Arabidopsis thaliana]
gi|23507781|gb|AAN38694.1| At3g04730/F7O18_22
[Arabidopsis thaliana]
Length = 236
Score = 145 bits (366), Expect = 4e-34
Identities = 66/72 (91%), Positives = 70/72 (96%)
Frame = -1
Query: 611 GMKDFMNESKLMDLLNSSDYVPTYEDKDGDWMLVGDVPWEMFVESCKRLRIMKGKEAIGL 432
GMKDFMNESKL+DLLN SDYVPTYEDKDGDWMLVGDVPWEMFV+SCKR+RIMKG EAIGL
Sbjct: 165 GMKDFMNESKLIDLLNGSDYVPTYEDKDGDWMLVGDVPWEMFVDSCKRIRIMKGSEAIGL 224
Query: 431 APRAMEKCKNRS 396
APRA+EKCKNRS
Sbjct: 225 APRALEKCKNRS 236
>gb|AAM96891.1| auxin-responsive protein IAA1; MjAux/IAA1 [Mirabilis jalapa]
Length = 194
Score = 144 bits (364), Expect = 7e-34
Identities = 67/71 (94%), Positives = 68/71 (95%)
Frame = -1
Query: 611 GMKDFMNESKLMDLLNSSDYVPTYEDKDGDWMLVGDVPWEMFVESCKRLRIMKGKEAIGL 432
GM DFMNESKLMDL+N SDYVPTYEDKDGDWMLVGDVPWEMFVESCKRLRIMKGKEA GL
Sbjct: 123 GMIDFMNESKLMDLINGSDYVPTYEDKDGDWMLVGDVPWEMFVESCKRLRIMKGKEAAGL 182
Query: 431 APRAMEKCKNR 399
APRAMEKCKNR
Sbjct: 183 APRAMEKCKNR 193
>gb|AAN16886.1| Aux/IAA1 [Mirabilis jalapa]
Length = 97
Score = 144 bits (364), Expect = 7e-34
Identities = 67/71 (94%), Positives = 68/71 (95%)
Frame = -1
Query: 611 GMKDFMNESKLMDLLNSSDYVPTYEDKDGDWMLVGDVPWEMFVESCKRLRIMKGKEAIGL 432
GM DFMNESKLMDL+N SDYVPTYEDKDGDWMLVGDVPWEMFVESCKRLRIMKGKEA GL
Sbjct: 26 GMIDFMNESKLMDLINGSDYVPTYEDKDGDWMLVGDVPWEMFVESCKRLRIMKGKEAAGL 85
Query: 431 APRAMEKCKNR 399
APRAMEKCKNR
Sbjct: 86 APRAMEKCKNR 96
>gb|AAD32147.1|AF123509_1 Nt-iaa4.1 deduced protein [Nicotiana tabacum]
Length = 220
Score = 144 bits (362), Expect = 1e-33
Identities = 66/72 (91%), Positives = 69/72 (95%)
Frame = -1
Query: 611 GMKDFMNESKLMDLLNSSDYVPTYEDKDGDWMLVGDVPWEMFVESCKRLRIMKGKEAIGL 432
G KDFMNESKL+DLLN SDYVPTYEDKDGDWMLVGDVPWEMFV+SCKRLRIMKG EAIGL
Sbjct: 149 GFKDFMNESKLIDLLNGSDYVPTYEDKDGDWMLVGDVPWEMFVDSCKRLRIMKGSEAIGL 208
Query: 431 APRAMEKCKNRS 396
APRA+EKCKNRS
Sbjct: 209 APRAVEKCKNRS 220
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 487,844,100
Number of Sequences: 1393205
Number of extensions: 10057875
Number of successful extensions: 21065
Number of sequences better than 10.0: 175
Number of HSP's better than 10.0 without gapping: 20537
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21053
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24568846532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)