Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
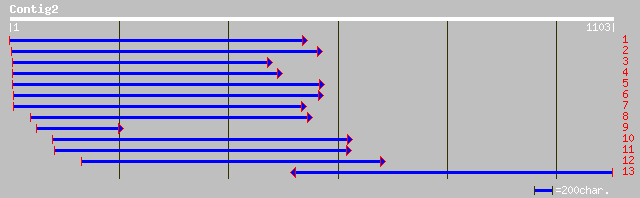
Query= KMC012346A_C02 KMC012346A_c02
(1096 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF64167.1|AF240462_1 plastid-specific ribosomal protein 2 pr... 277 2e-73
gb|AAL49922.1| unknown protein [Arabidopsis thaliana] gi|2213672... 275 9e-73
ref|NP_566958.2| RRM-containing protein; protein id: At3g52150.1... 268 8e-71
pir||T49094 hypothetical protein F4F15.260 - Arabidopsis thalian... 257 3e-67
gb|AAM11915.1| plastid-specific ribosomal protein 2 precursor [D... 159 7e-38
>gb|AAF64167.1|AF240462_1 plastid-specific ribosomal protein 2 precursor [Spinacia oleracea]
Length = 260
Score = 277 bits (708), Expect = 2e-73
Identities = 144/229 (62%), Positives = 177/229 (76%), Gaps = 1/229 (0%)
Frame = -3
Query: 1028 NLLSLTTPSSSTTSNLQPRHLRLAPSL-ARPLRLRRPFAVAEQAAATQSEADRRLYVGNI 852
NL+ L + S+ L+ R P L R + + + ++++ E RRLYVGNI
Sbjct: 32 NLVQLQSFSNGFGLKLKTRVSTNPPLLKVRAVVTEETSSSSTASSSSDGEGARRLYVGNI 91
Query: 851 PRTLTNDELANIVQEHAAVEKAEVMYDKYSGRSRRFAFVTVKTVEDANAAIEKLNGTQIG 672
PR L NDEL IV+EH A+E AEVMYDKYSGRSRRF FVT+KTVEDANA IEKLN T+IG
Sbjct: 92 PRNLNNDELRTIVEEHGAIEIAEVMYDKYSGRSRRFGFVTMKTVEDANAVIEKLNDTEIG 151
Query: 671 GREIKVNVTEKPLTTVDFPLVQAEESEYVDSPYKVYVGNLAKTVTSDMLKNFFSEKGKVL 492
GR+IKVN+TEKPL +D QAE+S++V+SPYKVY+GNLAKTVT+++LK+FFSEKGKVL
Sbjct: 152 GRKIKVNITEKPLEGMDIATTQAEDSQFVESPYKVYIGNLAKTVTNELLKDFFSEKGKVL 211
Query: 491 SAKVSRVPGTSKSSGFGFVTFSSDEDVEAAISSFNNAPLEGQKIRVNKA 345
AKV R PGTSKS+GFGFV+FSS+E+VEAAI + NN+ LEGQKIRVNKA
Sbjct: 212 GAKVQRTPGTSKSNGFGFVSFSSEEEVEAAIQALNNSVLEGQKIRVNKA 260
>gb|AAL49922.1| unknown protein [Arabidopsis thaliana] gi|22136722|gb|AAM91680.1|
unknown protein [Arabidopsis thaliana]
Length = 253
Score = 275 bits (703), Expect = 9e-73
Identities = 143/213 (67%), Positives = 174/213 (81%), Gaps = 7/213 (3%)
Frame = -3
Query: 962 LAPSLARPLRLRR-PFAVAEQ-----AAATQSEADRRLYVGNIPRTLTNDELANIVQEHA 801
LA + +R R R P+AV E A SEA RR+Y+GNIPRT+TN++L +V+EH
Sbjct: 41 LAGTFSRSSRTRFIPYAVTETEEKPAALDPSSEAARRVYIGNIPRTVTNEQLTKLVEEHG 100
Query: 800 AVEKAEVMYDKYSGRSRRFAFVTVKTVEDANAAIEKLNGTQIGGREIKVNVTEKPL-TTV 624
AVEK +VMYDKYSGRSRRF F T+K+VEDANA +EKLNG + GREIKVN+TEKP+ ++
Sbjct: 101 AVEKVQVMYDKYSGRSRRFGFATMKSVEDANAVVEKLNGNTVEGREIKVNITEKPIASSP 160
Query: 623 DFPLVQAEESEYVDSPYKVYVGNLAKTVTSDMLKNFFSEKGKVLSAKVSRVPGTSKSSGF 444
D ++Q+E+S +VDSPYKVYVGNLAKTVT +ML+N FSEKGKV+SAKVSRVPGTSKS+GF
Sbjct: 161 DLSVLQSEDSAFVDSPYKVYVGNLAKTVTKEMLENLFSEKGKVVSAKVSRVPGTSKSTGF 220
Query: 443 GFVTFSSDEDVEAAISSFNNAPLEGQKIRVNKA 345
GFVTFSS+EDVEAAI + NN+ LEGQKIRVNKA
Sbjct: 221 GFVTFSSEEDVEAAIVALNNSLLEGQKIRVNKA 253
>ref|NP_566958.2| RRM-containing protein; protein id: At3g52150.1 [Arabidopsis
thaliana]
Length = 260
Score = 268 bits (686), Expect = 8e-71
Identities = 143/220 (65%), Positives = 174/220 (79%), Gaps = 14/220 (6%)
Frame = -3
Query: 962 LAPSLARPLRLRR-PFAVAEQ-----AAATQSEADRRLYVGNIPRTLTNDELANIVQEHA 801
LA + +R R R P+AV E A SEA RR+Y+GNIPRT+TN++L +V+EH
Sbjct: 41 LAGTFSRSSRTRFIPYAVTETEEKPAALDPSSEAARRVYIGNIPRTVTNEQLTKLVEEHG 100
Query: 800 AVEKAEVMYDKYSGRSRRFAFVTVKTVEDANAAIEKLNGTQ-------IGGREIKVNVTE 642
AVEK +VMYDKYSGRSRRF F T+K+VEDANA +EKLNG + GREIKVN+TE
Sbjct: 101 AVEKVQVMYDKYSGRSRRFGFATMKSVEDANAVVEKLNGNSLFLVSQTVEGREIKVNITE 160
Query: 641 KPL-TTVDFPLVQAEESEYVDSPYKVYVGNLAKTVTSDMLKNFFSEKGKVLSAKVSRVPG 465
KP+ ++ D ++Q+E+S +VDSPYKVYVGNLAKTVT +ML+N FSEKGKV+SAKVSRVPG
Sbjct: 161 KPIASSPDLSVLQSEDSAFVDSPYKVYVGNLAKTVTKEMLENLFSEKGKVVSAKVSRVPG 220
Query: 464 TSKSSGFGFVTFSSDEDVEAAISSFNNAPLEGQKIRVNKA 345
TSKS+GFGFVTFSS+EDVEAAI + NN+ LEGQKIRVNKA
Sbjct: 221 TSKSTGFGFVTFSSEEDVEAAIVALNNSLLEGQKIRVNKA 260
>pir||T49094 hypothetical protein F4F15.260 - Arabidopsis thaliana
gi|4678944|emb|CAB41335.1| putative protein [Arabidopsis
thaliana]
Length = 546
Score = 257 bits (656), Expect = 3e-67
Identities = 142/230 (61%), Positives = 173/230 (74%), Gaps = 14/230 (6%)
Frame = -3
Query: 962 LAPSLARPLRLRR-PFAVAEQ-----AAATQSEADRRLYVGNIPRTLTNDELANIVQEHA 801
LA + +R R R P+AV E A SEA RR+Y+GNIPRT+TN++L +V+EH
Sbjct: 41 LAGTFSRSSRTRFIPYAVTETEEKPAALDPSSEAARRVYIGNIPRTVTNEQLTKLVEEHG 100
Query: 800 AVEKAEVMYDKYSGRSRRFAFVTVKTVEDANAAIEKLNGTQ-------IGGREIKVNVTE 642
AVEK VMYDKYSGRSRRF F T+K+VEDANA +EKLNG + GREIKVN+TE
Sbjct: 101 AVEK--VMYDKYSGRSRRFGFATMKSVEDANAVVEKLNGNSLFLVSQTVEGREIKVNITE 158
Query: 641 KPL-TTVDFPLVQAEESEYVDSPYKVYVGNLAKTVTSDMLKNFFSEKGKVLSAKVSRVPG 465
KP+ ++ D ++Q+E+S +VDSPYKVYVGNLAKTVT +ML+N FSEKGKV+SAKVSRVPG
Sbjct: 159 KPIASSPDLSVLQSEDSAFVDSPYKVYVGNLAKTVTKEMLENLFSEKGKVVSAKVSRVPG 218
Query: 464 TSKSSGFGFVTFSSDEDVEAAISSFNNAPLEGQKIRVNKA*RN*QCCRAC 315
TSKS+GFGFVTFSS+EDVEAAI + NN+ LEGQKIR+ CC C
Sbjct: 219 TSKSTGFGFVTFSSEEDVEAAIVALNNSLLEGQKIRL-------FCCLLC 261
>gb|AAM11915.1| plastid-specific ribosomal protein 2 precursor [Deschampsia
antarctica]
Length = 114
Score = 159 bits (402), Expect = 7e-38
Identities = 82/115 (71%), Positives = 94/115 (81%)
Frame = -3
Query: 689 NGTQIGGREIKVNVTEKPLTTVDFPLVQAEESEYVDSPYKVYVGNLAKTVTSDMLKNFFS 510
N T++GGR+IKVNVTE L +D P E +VDS YKVYVGNLAKTVT ++LKNFFS
Sbjct: 1 NETEVGGRKIKVNVTESFLPNID-PSAPEAEPAFVDSQYKVYVGNLAKTVTMEVLKNFFS 59
Query: 509 EKGKVLSAKVSRVPGTSKSSGFGFVTFSSDEDVEAAISSFNNAPLEGQKIRVNKA 345
EKG+VLSA VSRVPGT KS GFGFVTFSSDE+VEAA+S+FNN LEGQ IRVN+A
Sbjct: 60 EKGEVLSATVSRVPGTPKSKGFGFVTFSSDEEVEAAVSTFNNTELEGQAIRVNRA 114
Score = 52.8 bits (125), Expect = 1e-05
Identities = 28/87 (32%), Positives = 47/87 (53%)
Frame = -3
Query: 911 AEQAAATQSEADRRLYVGNIPRTLTNDELANIVQEHAAVEKAEVMYDKYSGRSRRFAFVT 732
A +A ++ ++YVGN+ +T+T + L N E V A V + +S+ F FVT
Sbjct: 26 APEAEPAFVDSQYKVYVGNLAKTVTMEVLKNFFSEKGEVLSATVSRVPGTPKSKGFGFVT 85
Query: 731 VKTVEDANAAIEKLNGTQIGGREIKVN 651
+ E+ AA+ N T++ G+ I+VN
Sbjct: 86 FSSDEEVEAAVSTFNNTELEGQAIRVN 112
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 935,046,914
Number of Sequences: 1393205
Number of extensions: 21075240
Number of successful extensions: 70183
Number of sequences better than 10.0: 2340
Number of HSP's better than 10.0 without gapping: 61622
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 68749
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 65614730658
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)