Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
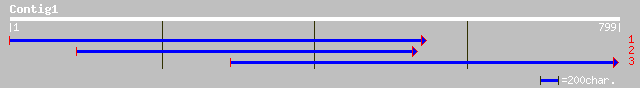
Query= KMC012313A_C01 KMC012313A_c01
(782 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_187542.1| putative alpha-hemolysin; protein id: At3g09310... 140 2e-32
ref|NP_283365.1| hypothetical protein NMA0549 [Neisseria meningi... 83 5e-15
ref|NP_274900.1| conserved hypothetical protein [Neisseria menin... 83 5e-15
ref|NP_624299.1| conserved hypothetical protein [Thermoanaerobac... 80 4e-14
ref|NP_229261.1| conserved hypothetical protein [Thermotoga mari... 78 2e-13
>ref|NP_187542.1| putative alpha-hemolysin; protein id: At3g09310.1 [Arabidopsis
thaliana] gi|14286021|sp|Q9SR32|Y331_ARATH Hypothetical
protein At3g09310 gi|6478923|gb|AAF14028.1|AC011436_12
putative alpha-hemolysin [Arabidopsis thaliana]
Length = 90
Score = 140 bits (354), Expect = 2e-32
Identities = 62/76 (81%), Positives = 68/76 (88%)
Frame = -3
Query: 738 MLRFYKREISPVLPTSCRYVPTCSEYSMEAYKRYGVVKGTVLTAWRLCRCNPLGGYGYDP 559
MLRFYKREISPVLP SCRYVPTCSEYSMEAYK+YGV+KGTVLT WRLCRCNPLGG G+DP
Sbjct: 1 MLRFYKREISPVLPRSCRYVPTCSEYSMEAYKKYGVLKGTVLTTWRLCRCNPLGGSGFDP 60
Query: 558 PRWFGEASPREEELDD 511
PRWFGE+ + E D+
Sbjct: 61 PRWFGESGIKLREGDE 76
>ref|NP_283365.1| hypothetical protein NMA0549 [Neisseria meningitidis Z2491]
gi|11279382|pir||E81973 hypothetical protein NMA0549
[imported] - Neisseria meningitidis (strain Z2491
serogroup A) gi|7379288|emb|CAB83842.1| hypothetical
protein NMA0549 [Neisseria meningitidis Z2491]
Length = 96
Score = 82.8 bits (203), Expect = 5e-15
Identities = 34/65 (52%), Positives = 48/65 (73%)
Frame = -3
Query: 753 KVALSMLRFYKREISPVLPTSCRYVPTCSEYSMEAYKRYGVVKGTVLTAWRLCRCNPLGG 574
K+ L ++RFY+ ISP++P CRY PTCS+Y++EA K+YG KG L R+ RC+P GG
Sbjct: 30 KLLLGLIRFYQYCISPLIPPRCRYTPTCSQYAVEAVKKYGAFKGGRLAIKRIARCHPFGG 89
Query: 573 YGYDP 559
+G+DP
Sbjct: 90 HGHDP 94
>ref|NP_274900.1| conserved hypothetical protein [Neisseria meningitidis MC58]
gi|14286015|sp|Q9JW47|Y549_NEIMA Hypothetical protein
NMA0549/NMB1906 gi|11279383|pir||A81028 conserved
hypothetical protein NMB1906 [imported] - Neisseria
meningitidis (strain MC58 serogroup B)
gi|7227163|gb|AAF42236.1| conserved hypothetical protein
[Neisseria meningitidis MC58]
Length = 73
Score = 82.8 bits (203), Expect = 5e-15
Identities = 34/65 (52%), Positives = 48/65 (73%)
Frame = -3
Query: 753 KVALSMLRFYKREISPVLPTSCRYVPTCSEYSMEAYKRYGVVKGTVLTAWRLCRCNPLGG 574
K+ L ++RFY+ ISP++P CRY PTCS+Y++EA K+YG KG L R+ RC+P GG
Sbjct: 7 KLLLGLIRFYQYCISPLIPPRCRYTPTCSQYAVEAVKKYGAFKGGRLAIKRIARCHPFGG 66
Query: 573 YGYDP 559
+G+DP
Sbjct: 67 HGHDP 71
>ref|NP_624299.1| conserved hypothetical protein [Thermoanaerobacter tengcongensis]
gi|25009634|sp|Q8R6K5|YS00_THETN Hypothetical protein
TTE2800 gi|20517808|gb|AAM25903.1| conserved
hypothetical protein [Thermoanaerobacter tengcongensis]
Length = 69
Score = 79.7 bits (195), Expect = 4e-14
Identities = 30/60 (50%), Positives = 46/60 (76%)
Frame = -3
Query: 738 MLRFYKREISPVLPTSCRYVPTCSEYSMEAYKRYGVVKGTVLTAWRLCRCNPLGGYGYDP 559
++R Y++ ISP+ P +CR+ PTCS+YS+EA +YG++KG +++ WR+ RCNP GYDP
Sbjct: 8 LIRLYQKYISPMKPRTCRFYPTCSQYSIEAISKYGLLKGGLMSIWRILRCNPFNPGGYDP 67
>ref|NP_229261.1| conserved hypothetical protein [Thermotoga maritima]
gi|14286076|sp|Q9X1H3|YE62_THEMA Hypothetical protein
TM1462 gi|7448119|pir||G72251 conserved hypothetical
protein - Thermotoga maritima (strain MSB8)
gi|4982027|gb|AAD36530.1|AE001797_10 conserved
hypothetical protein [Thermotoga maritima]
Length = 81
Score = 77.8 bits (190), Expect = 2e-13
Identities = 32/65 (49%), Positives = 48/65 (73%)
Frame = -3
Query: 753 KVALSMLRFYKREISPVLPTSCRYVPTCSEYSMEAYKRYGVVKGTVLTAWRLCRCNPLGG 574
K+ + ++RFY+R ISP+ P +CR+ PTCS Y ++A +++G++KGT L R+ RCNPL
Sbjct: 3 KLLIMLIRFYQRYISPLKPPTCRFTPTCSNYFIQALEKHGLLKGTFLGLRRILRCNPLSK 62
Query: 573 YGYDP 559
GYDP
Sbjct: 63 GGYDP 67
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 702,652,080
Number of Sequences: 1393205
Number of extensions: 16312571
Number of successful extensions: 42285
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 39909
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 42242
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 38935490438
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)