Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC012184A_C01 KMC012184A_c01
(601 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
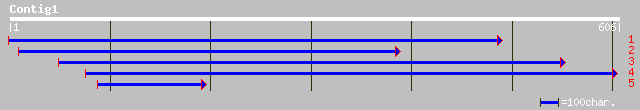
dbj|BAB60910.1| OSJNBa0054L14.2 [Oryza sativa (japonica cultivar... 108 7e-23
ref|NP_192449.1| putative protein; protein id: At4g05400.1 [Arab... 104 8e-22
ref|NP_193846.1| hypothetical protein; protein id: At4g21140.1 [... 59 4e-08
ref|XP_210569.1| similar to mitochondrial voltage dependent anio... 33 3.7
gb|AAL89810.1| Myb [Giardia intestinalis] gi|29249621|gb|EAA4112... 33 3.7
>dbj|BAB60910.1| OSJNBa0054L14.2 [Oryza sativa (japonica cultivar-group)]
gi|20804500|dbj|BAB92195.1| P0520B06.19 [Oryza sativa
(japonica cultivar-group)]
Length = 258
Score = 108 bits (269), Expect = 7e-23
Identities = 51/77 (66%), Positives = 65/77 (84%)
Frame = -2
Query: 600 VLMRKTRARQAAETNLLRMKKEAIEALPPHLKEAALVPDLAPFPANRFMATLTPPIPGYI 421
VLMR+ RARQ AE++LL +KKEAI ALP L+ AA++PD+ PFPANR+MATLTPPI GYI
Sbjct: 182 VLMRRHRARQTAESSLLSLKKEAIAALPEKLRAAAMIPDMTPFPANRYMATLTPPIEGYI 241
Query: 420 DKIREAANKISGREKIR 370
+K+R+AA K S +EK+R
Sbjct: 242 EKVRDAAKKYSVKEKLR 258
>ref|NP_192449.1| putative protein; protein id: At4g05400.1 [Arabidopsis thaliana]
gi|25341978|pir||H85067 hypothetical protein AT4g05400
[imported] - Arabidopsis thaliana
gi|7267300|emb|CAB81082.1| putative protein [Arabidopsis
thaliana]
Length = 250
Score = 104 bits (260), Expect = 8e-22
Identities = 52/77 (67%), Positives = 63/77 (81%)
Frame = -2
Query: 600 VLMRKTRARQAAETNLLRMKKEAIEALPPHLKEAALVPDLAPFPANRFMATLTPPIPGYI 421
VLMR+ R R+AAET LL +KK AIEALP +LK+AAL PDL PFPANR MATLTPPI GY+
Sbjct: 174 VLMREHRERRAAETALLNLKKSAIEALPENLKKAALEPDLTPFPANRGMATLTPPIEGYL 233
Query: 420 DKIREAANKISGREKIR 370
+K+ +AA K S +EK+R
Sbjct: 234 EKVMDAAKKSSSKEKLR 250
>ref|NP_193846.1| hypothetical protein; protein id: At4g21140.1 [Arabidopsis
thaliana] gi|7486599|pir||T04945 hypothetical protein
F7J7.80 - Arabidopsis thaliana
gi|2911071|emb|CAA17533.1| hypothetical protein
[Arabidopsis thaliana] gi|7268911|emb|CAB79114.1|
hypothetical protein [Arabidopsis thaliana]
Length = 228
Score = 59.3 bits (142), Expect = 4e-08
Identities = 28/47 (59%), Positives = 38/47 (80%)
Frame = -2
Query: 600 VLMRKTRARQAAETNLLRMKKEAIEALPPHLKEAALVPDLAPFPANR 460
VLMR+ RAR+ AE +L++M+ A+EALP +LK+AALV DL PFP +R
Sbjct: 166 VLMREHRARRVAEISLMKMRNAAVEALPENLKKAALVRDLTPFPVSR 212
>ref|XP_210569.1| similar to mitochondrial voltage dependent anion channel [Rattus
norvegicus] [Homo sapiens]
Length = 436
Score = 32.7 bits (73), Expect = 3.7
Identities = 14/31 (45%), Positives = 18/31 (57%)
Frame = +3
Query: 480 PSQGPKQPPSNVEARPQSPPSSSSKGSFLQP 572
P Q P+QPPS+ + PQSPP S+ P
Sbjct: 225 PQQPPQQPPSSNQQPPQSPPPPLSRRRLCHP 255
>gb|AAL89810.1| Myb [Giardia intestinalis] gi|29249621|gb|EAA41129.1|
GLP_306_56527_58119 [Giardia lamblia ATCC 50803]
Length = 530
Score = 32.7 bits (73), Expect = 3.7
Identities = 16/47 (34%), Positives = 26/47 (55%)
Frame = -1
Query: 286 CGPSRAVKIVLQNHNLEE*VLECQFFCVPFDSNDVKL*MANKFACLN 146
CG S+ + Q+H+L+E C + C +D+ DV L AN + L+
Sbjct: 237 CGQSKPI----QSHDLQEDTGHCSYSCTSYDTGDVSLPSANGSSTLS 279
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 536,864,550
Number of Sequences: 1393205
Number of extensions: 11799119
Number of successful extensions: 37674
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 34632
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37528
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23426109484
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)