Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC012071A_C01 KMC012071A_c01
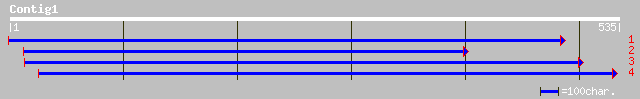
(531 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN86619.1| succinyl-CoA ligase alpha subunit [Lycopersicon e... 170 2e-64
ref|NP_197716.1| succinyl-CoA synthetase, alpha subunit; protein... 162 3e-62
gb|AAM65450.1| succinyl-CoA synthetase, alpha subunit [Arabidops... 162 3e-62
pir||S30579 succinate-CoA ligase (GDP-forming) (EC 6.2.1.4) alph... 160 1e-61
ref|NP_196447.1| succinyl-CoA-ligase alpha subunit; protein id: ... 158 1e-60
>gb|AAN86619.1| succinyl-CoA ligase alpha subunit [Lycopersicon esculentum]
Length = 332
Score = 170 bits (430), Expect(2) = 2e-64
Identities = 83/89 (93%), Positives = 85/89 (95%)
Frame = +3
Query: 114 ASSPAVFVDKSTRVICQGITGKNGTFHTEQAIEYGTNMVGGVTPKKGGTEHLGLPVFNTV 293
A+ PAVFVDK+TRVICQGITGKNGTFHTEQAIEYGT MVGGVTPKKGGTEHLGLPVFNTV
Sbjct: 34 AAPPAVFVDKNTRVICQGITGKNGTFHTEQAIEYGTKMVGGVTPKKGGTEHLGLPVFNTV 93
Query: 294 AEAKAETKANASVIYVPPPFAAKAIMEGL 380
EAKAETKANASVIYVPPPFAA AIMEGL
Sbjct: 94 EEAKAETKANASVIYVPPPFAAAAIMEGL 122
Score = 97.4 bits (241), Expect(2) = 2e-64
Identities = 45/51 (88%), Positives = 49/51 (95%)
Frame = +1
Query: 379 LEAELDLVVCITEGIPQHDMVRVKAAINRQSKTRLIGPNWPGIIKPGECKI 531
LEAELDL+VCITEGIPQHDMVRVKAA+ +QS+TRLIGPN PGIIKPGECKI
Sbjct: 122 LEAELDLIVCITEGIPQHDMVRVKAALKKQSRTRLIGPNCPGIIKPGECKI 172
>ref|NP_197716.1| succinyl-CoA synthetase, alpha subunit; protein id: At5g23250.1,
supported by cDNA: 39462., supported by cDNA:
gi_20260189 [Arabidopsis thaliana]
gi|10177814|dbj|BAB11180.1| succinyl-CoA synthetase,
alpha subunit [Arabidopsis thaliana]
gi|20260190|gb|AAM12993.1| succinyl-CoA synthetase,
alpha subunit [Arabidopsis thaliana]
gi|21387045|gb|AAM47926.1| succinyl-CoA synthetase alpha
subunit [Arabidopsis thaliana]
Length = 341
Score = 162 bits (411), Expect(2) = 3e-62
Identities = 83/96 (86%), Positives = 84/96 (87%), Gaps = 1/96 (1%)
Frame = +3
Query: 96 RHLSATASSPA-VFVDKSTRVICQGITGKNGTFHTEQAIEYGTNMVGGVTPKKGGTEHLG 272
R T PA VFVDK+TRVICQGITGKNGTFHTEQAIEYGT MV GVTPKKGGTEHLG
Sbjct: 35 RSFGTTPPPPAAVFVDKNTRVICQGITGKNGTFHTEQAIEYGTKMVAGVTPKKGGTEHLG 94
Query: 273 LPVFNTVAEAKAETKANASVIYVPPPFAAKAIMEGL 380
LPVFNTVAEAKAETKANASVIYVP PFAA AIMEGL
Sbjct: 95 LPVFNTVAEAKAETKANASVIYVPAPFAAAAIMEGL 130
Score = 97.4 bits (241), Expect(2) = 3e-62
Identities = 46/51 (90%), Positives = 48/51 (93%)
Frame = +1
Query: 379 LEAELDLVVCITEGIPQHDMVRVKAAINRQSKTRLIGPNWPGIIKPGECKI 531
L AELDL+VCITEGIPQHDMVRVKAA+N QSKTRLIGPN PGIIKPGECKI
Sbjct: 130 LAAELDLIVCITEGIPQHDMVRVKAALNSQSKTRLIGPNCPGIIKPGECKI 180
>gb|AAM65450.1| succinyl-CoA synthetase, alpha subunit [Arabidopsis thaliana]
Length = 341
Score = 162 bits (411), Expect(2) = 3e-62
Identities = 83/96 (86%), Positives = 84/96 (87%), Gaps = 1/96 (1%)
Frame = +3
Query: 96 RHLSATASSPA-VFVDKSTRVICQGITGKNGTFHTEQAIEYGTNMVGGVTPKKGGTEHLG 272
R T PA VFVDK+TRVICQGITGKNGTFHTEQAIEYGT MV GVTPKKGGTEHLG
Sbjct: 35 RSFGTTPPPPAAVFVDKNTRVICQGITGKNGTFHTEQAIEYGTKMVAGVTPKKGGTEHLG 94
Query: 273 LPVFNTVAEAKAETKANASVIYVPPPFAAKAIMEGL 380
LPVFNTVAEAKAETKANASVIYVP PFAA AIMEGL
Sbjct: 95 LPVFNTVAEAKAETKANASVIYVPAPFAAAAIMEGL 130
Score = 97.4 bits (241), Expect(2) = 3e-62
Identities = 46/51 (90%), Positives = 48/51 (93%)
Frame = +1
Query: 379 LEAELDLVVCITEGIPQHDMVRVKAAINRQSKTRLIGPNWPGIIKPGECKI 531
L AELDL+VCITEGIPQHDMVRVKAA+N QSKTRLIGPN PGIIKPGECKI
Sbjct: 130 LAAELDLIVCITEGIPQHDMVRVKAALNSQSKTRLIGPNCPGIIKPGECKI 180
>pir||S30579 succinate-CoA ligase (GDP-forming) (EC 6.2.1.4) alpha chain -
Arabidopsis thaliana (fragment) gi|16510|emb|CAA48891.1|
succinate--CoA ligase (GDP-forming) [Arabidopsis
thaliana]
Length = 345
Score = 160 bits (406), Expect(2) = 1e-61
Identities = 79/84 (94%), Positives = 80/84 (95%)
Frame = +3
Query: 129 VFVDKSTRVICQGITGKNGTFHTEQAIEYGTNMVGGVTPKKGGTEHLGLPVFNTVAEAKA 308
VFVDK+TRVICQGITGKNGTFHTEQAIEYGT MV GVTPKKGGTEHLGLPVFNTVAEAKA
Sbjct: 51 VFVDKNTRVICQGITGKNGTFHTEQAIEYGTKMVAGVTPKKGGTEHLGLPVFNTVAEAKA 110
Query: 309 ETKANASVIYVPPPFAAKAIMEGL 380
ETKANASVIYVP PFAA AIMEGL
Sbjct: 111 ETKANASVIYVPAPFAAAAIMEGL 134
Score = 97.4 bits (241), Expect(2) = 1e-61
Identities = 46/51 (90%), Positives = 48/51 (93%)
Frame = +1
Query: 379 LEAELDLVVCITEGIPQHDMVRVKAAINRQSKTRLIGPNWPGIIKPGECKI 531
L AELDL+VCITEGIPQHDMVRVKAA+N QSKTRLIGPN PGIIKPGECKI
Sbjct: 134 LAAELDLIVCITEGIPQHDMVRVKAALNSQSKTRLIGPNCPGIIKPGECKI 184
>ref|NP_196447.1| succinyl-CoA-ligase alpha subunit; protein id: At5g08300.1,
supported by cDNA: 10292., supported by cDNA:
gi_18252214 [Arabidopsis thaliana]
gi|19884199|sp|P53586|SUCA_ARATH Succinyl-CoA ligase
[GDP-forming] alpha-chain, mitochondrial precursor
(Succinyl-CoA synthetase, alpha chain) (SCS-alpha)
gi|11272000|pir||T51816 succinate-CoA ligase
(GDP-forming) (EC 6.2.1.4) alpha chain [imported] -
Arabidopsis thaliana gi|3660467|emb|CAA05023.1|
succinyl-CoA-ligase alpha subunit [Arabidopsis thaliana]
gi|10178272|emb|CAC08330.1| succinyl-CoA-ligase alpha
subunit [Arabidopsis thaliana]
gi|18252215|gb|AAL61940.1| succinyl-CoA-ligase alpha
subunit [Arabidopsis thaliana]
gi|21387057|gb|AAM47932.1| succinyl-CoA-ligase alpha
subunit [Arabidopsis thaliana]
gi|21536557|gb|AAM60889.1| succinyl-CoA-ligase alpha
subunit [Arabidopsis thaliana]
Length = 347
Score = 158 bits (399), Expect(2) = 1e-60
Identities = 76/85 (89%), Positives = 81/85 (94%)
Frame = +3
Query: 126 AVFVDKSTRVICQGITGKNGTFHTEQAIEYGTNMVGGVTPKKGGTEHLGLPVFNTVAEAK 305
AVFVDK+TRV+CQGITGKNGTFHTEQAIEYGT MV GVTPKKGGTEHLGLPVFN+VAEAK
Sbjct: 51 AVFVDKNTRVLCQGITGKNGTFHTEQAIEYGTKMVAGVTPKKGGTEHLGLPVFNSVAEAK 110
Query: 306 AETKANASVIYVPPPFAAKAIMEGL 380
A+TKANASVIYVP PFAA AIMEG+
Sbjct: 111 ADTKANASVIYVPAPFAAAAIMEGI 135
Score = 96.7 bits (239), Expect(2) = 1e-60
Identities = 45/51 (88%), Positives = 48/51 (93%)
Frame = +1
Query: 379 LEAELDLVVCITEGIPQHDMVRVKAAINRQSKTRLIGPNWPGIIKPGECKI 531
+EAELDL+VCITEGIPQHDMVRVK A+N QSKTRLIGPN PGIIKPGECKI
Sbjct: 135 IEAELDLIVCITEGIPQHDMVRVKHALNSQSKTRLIGPNCPGIIKPGECKI 185
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 528,718,919
Number of Sequences: 1393205
Number of extensions: 13017967
Number of successful extensions: 55863
Number of sequences better than 10.0: 320
Number of HSP's better than 10.0 without gapping: 48298
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 54407
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17596710992
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)