Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011986A_C01 KMC011986A_c01
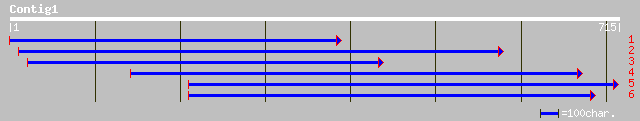
(714 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_565175.1| RRM-containing protein; protein id: At1g78260.1... 74 2e-12
pir||H96811 protein F3F9.20 [imported] - Arabidopsis thaliana gi... 69 6e-11
pir||B86356 T16E15.6 protein - Arabidopsis thaliana gi|9392682|g... 63 4e-09
ref|NP_564169.1| glycine-rich RNA-binding protein, putative; pro... 45 8e-04
ref|NP_649286.1| CG12976-PA [Drosophila melanogaster] gi|7296394... 45 0.001
>ref|NP_565175.1| RRM-containing protein; protein id: At1g78260.1, supported by cDNA:
123429. [Arabidopsis thaliana]
gi|21537200|gb|AAM61541.1| RNA recognition
motif-containing protein SEB-4 [Arabidopsis thaliana]
Length = 287
Score = 73.9 bits (180), Expect = 2e-12
Identities = 53/150 (35%), Positives = 76/150 (50%), Gaps = 28/150 (18%)
Frame = -3
Query: 709 VLYPPYGYPTYTPDHYGYHQAAMYNPQIHHQQPQYY--HQQMYGAASSS----------T 566
++YP YGY TY PD +GYHQ A+YN Q+ QQ QYY QQ+YG ++S +
Sbjct: 137 LMYPSYGY-TYNPDQFGYHQ-ALYNTQL--QQAQYYQQQQQLYGGGATSPSSSGATIMPS 192
Query: 565 PYYYGYSVQAAPRGTFS---------TPQAHRMP--AGPSYLYYPTT-----PIEGSFSA 434
PYYYGYS+Q APR + PQ H + PS+L YP+ P++G S+
Sbjct: 193 PYYYGYSLQ-APRVPYQHHHHLPQPYNPQQHHHQRFSSPSFLVYPSNSSFAPPLQGVLSS 251
Query: 433 YRPIQQQPVIRQPSPSPSDSQTQQRTSSET 344
+ P + + + + T+S T
Sbjct: 252 STESEAVPQQVPAAEGEATTTAPEITTSNT 281
>pir||H96811 protein F3F9.20 [imported] - Arabidopsis thaliana
gi|8052542|gb|AAF71806.1|AC013430_15 F3F9.20
[Arabidopsis thaliana]
Length = 302
Score = 68.9 bits (167), Expect = 6e-11
Identities = 53/164 (32%), Positives = 76/164 (46%), Gaps = 42/164 (25%)
Frame = -3
Query: 709 VLYPPYGYPTYTPDHYGYHQA--------------AMYNPQIHHQQPQYYHQQ--MYGAA 578
++YP YGY TY PD +GYHQ A+YN Q+ QQ QYY QQ +YG
Sbjct: 137 LMYPSYGY-TYNPDQFGYHQTSTMMMMMMIIYLPQALYNTQL--QQAQYYQQQQQLYGGG 193
Query: 577 SSS----------TPYYYGYSVQAAPRGTFS---------TPQAHRMP--AGPSYLYYPT 461
++S +PYYYGYS+Q APR + PQ H + PS+L YP+
Sbjct: 194 ATSPSSSGATIMPSPYYYGYSLQ-APRVPYQHHHHLPQPYNPQQHHHQRFSSPSFLVYPS 252
Query: 460 T-----PIEGSFSAYRPIQQQPVIRQPSPSPSDSQTQQRTSSET 344
P++G S+ + P + + + + T+S T
Sbjct: 253 NSSFAPPLQGVLSSSTESEAVPQQVPAAEGEATTTAPEITTSNT 296
>pir||B86356 T16E15.6 protein - Arabidopsis thaliana
gi|9392682|gb|AAF87259.1|AC068562_6 Contains similarity
to seb4D protein from Mus musculus gb|X75316 and
contains a RNA recognition PF|00076 motif. [Arabidopsis
thaliana]
Length = 291
Score = 63.2 bits (152), Expect = 4e-09
Identities = 48/142 (33%), Positives = 66/142 (45%), Gaps = 21/142 (14%)
Frame = -3
Query: 709 VLYPPYGYPTYTPDHYGYHQAAMYNPQIHHQQPQYYHQQMYGAASSS---------TPYY 557
++YP YGY TY + YGYHQA +YN Q+ QQ QYY QQMYG ++ +PYY
Sbjct: 137 LMYPSYGY-TYNSE-YGYHQA-LYNAQL--QQAQYYQQQMYGGGGATSPSSSNIMPSPYY 191
Query: 556 YGYSVQAAP------------RGTFSTPQAHRMPAGPSYLYYPTTPIEGSFSAYRPIQQQ 413
Y + P Q R+P+ SYL YP+ +E + P Q
Sbjct: 192 YLQAPSPRPYLHHQQHYQYHHHQQQQQQQQQRLPSASSYLIYPSN-VEAPTPSNVPTSQG 250
Query: 412 PVIRQPSPSPSDSQTQQRTSSE 347
P+ S ++S Q+ S E
Sbjct: 251 PL-----SSSTESHAPQQVSEE 267
>ref|NP_564169.1| glycine-rich RNA-binding protein, putative; protein id: At1g22335.1
[Arabidopsis thaliana]
Length = 139
Score = 45.4 bits (106), Expect = 8e-04
Identities = 36/121 (29%), Positives = 51/121 (41%), Gaps = 21/121 (17%)
Frame = -3
Query: 646 AMYNPQIHHQQPQYYHQQMYGAASSS---------TPYYYGYSVQAAP------------ 530
A+YN Q+ QQ QYY QQMYG ++ +PYYY + P
Sbjct: 3 ALYNAQL--QQAQYYQQQMYGGGGATSPSSSNIMPSPYYYLQAPSPRPYLHHQQHYQYHH 60
Query: 529 RGTFSTPQAHRMPAGPSYLYYPTTPIEGSFSAYRPIQQQPVIRQPSPSPSDSQTQQRTSS 350
Q R+P+ SYL YP+ +E + P Q P+ S ++S Q+ S
Sbjct: 61 HQQQQQQQQQRLPSASSYLIYPSN-VEAPTPSNVPTSQGPL-----SSSTESHAPQQVSE 114
Query: 349 E 347
E
Sbjct: 115 E 115
>ref|NP_649286.1| CG12976-PA [Drosophila melanogaster] gi|7296394|gb|AAF51682.1|
CG12976-PA [Drosophila melanogaster]
Length = 314
Score = 45.1 bits (105), Expect = 0.001
Identities = 37/121 (30%), Positives = 51/121 (41%), Gaps = 18/121 (14%)
Frame = -3
Query: 688 YP-TYTPDHYGYHQAAMYNPQIHHQQPQYY-------HQQMYGAASSST----PYYYGYS 545
YP ++ H+ +H P + H PQ Y H YG SSS+ P YY Y+
Sbjct: 5 YPASFYDQHHSHHHPQHLTPTMSHYPPQPYRSGYTGYHTYGYGYGSSSSASYMPNYYRYA 64
Query: 544 VQAAP------RGTFSTPQAHRMPAGPSYLYYPTTPIEGSFSAYRPIQQQPVIRQPSPSP 383
A+P +G F+ +P+ S L+YP P P QQ P +QP P
Sbjct: 65 PYASPHYSQHHQGMFTPAGQQIIPS--SVLHYPPRP--------HPYQQLPQQQQPHPQQ 114
Query: 382 S 380
S
Sbjct: 115 S 115
Score = 33.1 bits (74), Expect = 3.9
Identities = 41/174 (23%), Positives = 58/174 (32%), Gaps = 46/174 (26%)
Frame = -3
Query: 703 YPPYGYPTYTPDHYGYHQAA----------MYNPQIH--------------HQQPQY--- 605
Y PY P Y+ H G A Y P+ H QP Y
Sbjct: 63 YAPYASPHYSQHHQGMFTPAGQQIIPSSVLHYPPRPHPYQQLPQQQQPHPQQSQPGYEPP 122
Query: 604 ----------YHQQMYGAASSSTPYYYGYSVQAAPRGTFSTPQAHRMPAGPSYLYYPTTP 455
Y + +GA + TP+Y AP G STP +R PSYL P
Sbjct: 123 PPLPYSSSASYQSRGFGAFAGPTPHY-------APPGRSSTPAPNRTVLPPSYL----EP 171
Query: 454 IEGSFSAYRPIQQQPVIRQP---------SPSPSDSQTQQRTSSETSAGVVITS 320
+ + + + QQ + Q +P D + +E+ A + TS
Sbjct: 172 LRSYSAEQQHLSQQQHLSQQQLQHHPLPGTPKLEDPDVEVEAVAESPAQRLTTS 225
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 646,115,264
Number of Sequences: 1393205
Number of extensions: 14990035
Number of successful extensions: 55237
Number of sequences better than 10.0: 285
Number of HSP's better than 10.0 without gapping: 49994
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 54360
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32936043699
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)