Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
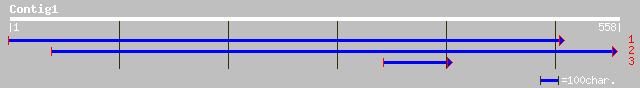
Query= KMC011969A_C01 KMC011969A_c01
(558 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_187476.1| unknown protein; protein id: At3g08640.1, suppo... 166 1e-40
ref|NP_187475.1| unknown protein; protein id: At3g08630.1 [Arabi... 160 1e-38
dbj|BAB85250.1| P0425G02.25 [Oryza sativa (japonica cultivar-gro... 159 2e-38
gb|AAM44363.1| hypothetical protein [Dictyostelium discoideum] g... 60 2e-08
sp|P04929|HRPX_PLALO HISTIDINE-RICH GLYCOPROTEIN PRECURSOR gi|72... 59 5e-08
>ref|NP_187476.1| unknown protein; protein id: At3g08640.1, supported by cDNA:
gi_19698960 [Arabidopsis thaliana]
gi|12322723|gb|AAG51347.1|AC012562_8 unknown protein;
33915-34928 [Arabidopsis thaliana]
gi|19698961|gb|AAL91216.1| unknown protein [Arabidopsis
thaliana] gi|22136296|gb|AAM91226.1| unknown protein
[Arabidopsis thaliana]
Length = 337
Score = 166 bits (421), Expect = 1e-40
Identities = 81/113 (71%), Positives = 90/113 (78%), Gaps = 1/113 (0%)
Frame = +1
Query: 223 AGGDGGDGVGRGGGGGG-GGGGDESKSDGGSGNSNDSSFGILGLFLNGWRSRVAADPQFP 399
AGG GG G GGG G GGGG GG + S +G +GLF+ GWRSRVAADPQFP
Sbjct: 60 AGGGGGGSTGNNGGGSGSGGGGGGFGGSGGEASEESSPWGPIGLFIQGWRSRVAADPQFP 119
Query: 400 FKVLMEELVGVTACVLGDMASRPNFGLNELDFVFSTLVVGAILNFTLMYLLAP 558
FKVLMEE+VG++ACVLGDMASRPNFGLNELDFVFSTLVVG+ILNF LMY+LAP
Sbjct: 120 FKVLMEEIVGLSACVLGDMASRPNFGLNELDFVFSTLVVGSILNFVLMYMLAP 172
>ref|NP_187475.1| unknown protein; protein id: At3g08630.1 [Arabidopsis thaliana]
gi|12322720|gb|AAG51344.1|AC012562_5 unknown protein;
31866-32885 [Arabidopsis thaliana]
Length = 339
Score = 160 bits (405), Expect = 1e-38
Identities = 83/112 (74%), Positives = 88/112 (78%)
Frame = +1
Query: 223 AGGDGGDGVGRGGGGGGGGGGDESKSDGGSGNSNDSSFGILGLFLNGWRSRVAADPQFPF 402
AGG GG +G GGG G GGG GGS S +G LGLF+ GWRSRVAAD QFPF
Sbjct: 60 AGGGGGGSIGNHGGGSGSGGGGGGY--GGSEEEESSPWGPLGLFIQGWRSRVAADSQFPF 117
Query: 403 KVLMEELVGVTACVLGDMASRPNFGLNELDFVFSTLVVGAILNFTLMYLLAP 558
KVLME LVGV+A VLGDMASRPNFGLNELDFVFSTLVVG+ILNFTLMYLLAP
Sbjct: 118 KVLMEMLVGVSANVLGDMASRPNFGLNELDFVFSTLVVGSILNFTLMYLLAP 169
>dbj|BAB85250.1| P0425G02.25 [Oryza sativa (japonica cultivar-group)]
gi|20161467|dbj|BAB90391.1| P0432B10.9 [Oryza sativa
(japonica cultivar-group)]
Length = 401
Score = 159 bits (402), Expect = 2e-38
Identities = 99/170 (58%), Positives = 112/170 (65%), Gaps = 6/170 (3%)
Frame = +1
Query: 67 SMATTALLRFSPLSGHSHALSFPS-SLPSPQTQNRSIYLTNPPRLPERLQLSFAGGDGGD 243
SMA TA +F P + +H S P S P + + S ++ L RL+ G G
Sbjct: 56 SMAFTAA-KFLPATAPTHLDSSPRLSPPRAGSLSFSPLSSSSSALLLRLRSPSPSGPSGP 114
Query: 244 GVGR-----GGGGGGGGGGDESKSDGGSGNSNDSSFGILGLFLNGWRSRVAADPQFPFKV 408
G GR GGGGG GD + S G SG GILG+FL GW +RVAADPQFPFKV
Sbjct: 115 G-GRLPPPPRSYGGGGGSGDAADSGGSSG-------GILGIFLAGWAARVAADPQFPFKV 166
Query: 409 LMEELVGVTACVLGDMASRPNFGLNELDFVFSTLVVGAILNFTLMYLLAP 558
LMEELVGV+ACVLGDMASRPNFGLNELDFVFSTLVVG+ILNF LMYLLAP
Sbjct: 167 LMEELVGVSACVLGDMASRPNFGLNELDFVFSTLVVGSILNFVLMYLLAP 216
>gb|AAM44363.1| hypothetical protein [Dictyostelium discoideum]
gi|28828387|gb|AAM09303.2| similar to Plasmodium
lophurae. Histidine-rich glycoprotein precursor
[Dictyostelium discoideum]
Length = 233
Score = 60.1 bits (144), Expect = 2e-08
Identities = 26/74 (35%), Positives = 29/74 (39%)
Frame = -3
Query: 451 HQEHKQSHLPTPPSKP*MEIVDQQQPLISIHSERDPKSQKMNHWNFHFHHHSYFHHHHHH 272
H H H P P P H P +H + H HHH + HHHHHH
Sbjct: 88 HHHHHHHHHPHHP---------HHHP----HHHHHPHHHHHHHHHHHHHHHHHHHHHHHH 134
Query: 271 RHHLHHAQPHHRRH 230
HH HH HH H
Sbjct: 135 HHHHHHHHHHHHHH 148
Score = 60.1 bits (144), Expect = 2e-08
Identities = 22/61 (36%), Positives = 33/61 (54%), Gaps = 4/61 (6%)
Frame = -3
Query: 400 MEIVDQQQPLISIHSERDPKSQKMN----HWNFHFHHHSYFHHHHHHRHHLHHAQPHHRR 233
+++ + P + H+ +P + + H + H HHH + HHHHHH HH HH PHH
Sbjct: 42 LDVNNPHNPNNNPHNPHNPNNNPHHPHHLHHHHHHHHHHHHHHHHHHHHHHHHHHPHHPH 101
Query: 232 H 230
H
Sbjct: 102 H 102
Score = 59.3 bits (142), Expect = 3e-08
Identities = 19/33 (57%), Positives = 22/33 (66%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
+H + H HHH + HHHHHH HH HH PHH H
Sbjct: 128 HHHHHHHHHHHHHHHHHHHHHHHHHHHPHHHPH 160
Score = 57.8 bits (138), Expect = 9e-08
Identities = 25/74 (33%), Positives = 28/74 (37%)
Frame = -3
Query: 451 HQEHKQSHLPTPPSKP*MEIVDQQQPLISIHSERDPKSQKMNHWNFHFHHHSYFHHHHHH 272
H H H P P P H +H + H HHH + HHHHHH
Sbjct: 85 HHHHHHHHHHHHPHHPHHHPHHHHHP----HHHHHHHHHHHHHHHHHHHHHHHHHHHHHH 140
Query: 271 RHHLHHAQPHHRRH 230
HH HH HH H
Sbjct: 141 HHHHHHHHHHHHHH 154
Score = 55.8 bits (133), Expect = 3e-07
Identities = 18/33 (54%), Positives = 21/33 (63%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
+H + H HHH + HHHHHH HH HH HH H
Sbjct: 125 HHHHHHHHHHHHHHHHHHHHHHHHHHHHHHPHH 157
Score = 55.5 bits (132), Expect = 4e-07
Identities = 18/33 (54%), Positives = 21/33 (63%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
+H + H HHH + HHHHHH HH H PHH H
Sbjct: 73 HHHHHHHHHHHHHHHHHHHHHHHHPHHPHHHPH 105
Score = 55.5 bits (132), Expect = 4e-07
Identities = 19/35 (54%), Positives = 22/35 (62%), Gaps = 2/35 (5%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHA--QPHHRRH 230
+H + H HHH + HHHHHH HH HH PHH H
Sbjct: 75 HHHHHHHHHHHHHHHHHHHHHHPHHPHHHPHHHHH 109
Score = 52.4 bits (124), Expect = 4e-06
Identities = 19/42 (45%), Positives = 22/42 (52%), Gaps = 9/42 (21%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHH---------LHHAQPHHRRH 230
+H + H HHH + HHHHHH HH HH PHH H
Sbjct: 74 HHHHHHHHHHHHHHHHHHHHHHHPHHPHHHPHHHHHPHHHHH 115
Score = 52.0 bits (123), Expect = 5e-06
Identities = 17/33 (51%), Positives = 20/33 (60%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
+H + H HHH + HHHHHH H HH PH H
Sbjct: 134 HHHHHHHHHHHHHHHHHHHHHPHHHPHPHPHPH 166
Score = 51.6 bits (122), Expect = 6e-06
Identities = 17/33 (51%), Positives = 20/33 (60%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
+H + H HHH + HHHHHH HH H PH H
Sbjct: 132 HHHHHHHHHHHHHHHHHHHHHHHPHHHPHPHPH 164
Score = 51.2 bits (121), Expect = 8e-06
Identities = 17/33 (51%), Positives = 20/33 (60%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
+H + H HHH + HHHHHH HH H PH H
Sbjct: 136 HHHHHHHHHHHHHHHHHHHPHHHPHPHPHPHPH 168
Score = 48.5 bits (114), Expect = 5e-05
Identities = 19/54 (35%), Positives = 23/54 (42%), Gaps = 9/54 (16%)
Frame = -3
Query: 364 IHSERDPKSQKMNHWNFHFHHHSYFH---------HHHHHRHHLHHAQPHHRRH 230
+H +H + H HHH + H HHHHH HH HH HH H
Sbjct: 70 LHHHHHHHHHHHHHHHHHHHHHHHHHHPHHPHHHPHHHHHPHHHHHHHHHHHHH 123
Score = 43.5 bits (101), Expect = 0.002
Identities = 15/33 (45%), Positives = 18/33 (54%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
+H + H HHH + HH HHH H H PH H
Sbjct: 140 HHHHHHHHHHHHHHHPHHHPHPHPHPHPHPHLH 172
Score = 36.6 bits (83), Expect = 0.21
Identities = 17/41 (41%), Positives = 19/41 (45%), Gaps = 8/41 (19%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRH-----HLH---HAQPHHRRH 230
+H + H HHH HH H H H HLH H PH H
Sbjct: 144 HHHHHHHHHHHPHHHPHPHPHPHPHPHLHPNPHPHPHPHPH 184
Score = 35.4 bits (80), Expect = 0.48
Identities = 23/86 (26%), Positives = 28/86 (31%)
Frame = -3
Query: 451 HQEHKQSHLPTPPSKP*MEIVDQQQPLISIHSERDPKSQKMNHWNFHFHHHSYFHHHHHH 272
H H H P P P P +H P H + H HHH + H H
Sbjct: 150 HHHHHPHHHPHPHPHP--------HPHPHLHPNPHPHPHPHPHPHPHPHHHPNPNPHPHP 201
Query: 271 RHHLHHAQPHHRRHLRRRKVAIFQAN 194
H H HH H + +Q N
Sbjct: 202 HPHPH----HHHHHQEASECLSYQGN 223
>sp|P04929|HRPX_PLALO HISTIDINE-RICH GLYCOPROTEIN PRECURSOR gi|72400|pir||KGZQHL
histidine-rich glycoprotein precursor - Plasmodium
lophurae gi|9999|emb|CAA25698.1| histidine-rich protein
[Plasmodium lophurae] gi|224316|prf||1101401A
protein,His rich
Length = 351
Score = 58.5 bits (140), Expect = 5e-08
Identities = 28/77 (36%), Positives = 32/77 (41%)
Frame = -3
Query: 460 KPCHQEHKQSHLPTPPSKP*MEIVDQQQPLISIHSERDPKSQKMNHWNFHFHHHSYFHHH 281
+P H+EH H P +P E P H P +H H HHH HHH
Sbjct: 71 EPHHEEHHHHH-PEEHHEPHHEEHHHHHPHPHHHHHHHPPHHH-HHLGHHHHHHHAAHHH 128
Query: 280 HHHRHHLHHAQPHHRRH 230
HH HH HH HH H
Sbjct: 129 HHEEHHHHHHAAHHHHH 145
Score = 53.9 bits (128), Expect = 1e-06
Identities = 25/76 (32%), Positives = 30/76 (38%), Gaps = 2/76 (2%)
Frame = -3
Query: 451 HQEHKQSHLPTPPSKP*MEIVDQQQPLISIHSERDPKSQKMNHWNFHFHHHSYFHHHHHH 272
H+EH H + +P H P+ H H HHH + HHHHHH
Sbjct: 58 HEEHHHHHPE-----------EHHEPHHEEHHHHHPEEHHEPHHEEHHHHHPHPHHHHHH 106
Query: 271 R--HHLHHAQPHHRRH 230
HH HH HH H
Sbjct: 107 HPPHHHHHLGHHHHHH 122
Score = 52.4 bits (124), Expect = 4e-06
Identities = 17/30 (56%), Positives = 20/30 (66%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHAQPHH 239
+H + H HHH + HHHHHH HH HH HH
Sbjct: 221 HHGHHHHHHHHHGHHHHHHHHHGHHHHHHH 250
Score = 52.4 bits (124), Expect = 4e-06
Identities = 17/33 (51%), Positives = 20/33 (60%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
+H + H HHH + HHH HH HH HH HH H
Sbjct: 227 HHHHHHGHHHHHHHHHGHHHHHHHHHDAHHHHH 259
Score = 51.6 bits (122), Expect = 6e-06
Identities = 17/32 (53%), Positives = 19/32 (59%)
Frame = -3
Query: 325 HWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
H + H HHH + HHHHHH H HH HH H
Sbjct: 224 HHHHHHHHHGHHHHHHHHHGHHHHHHHHHDAH 255
Score = 51.6 bits (122), Expect = 6e-06
Identities = 18/36 (50%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHH---RHHLHHAQPHHRRH 230
+H +H HH + HHHHHH HH HH PHH H
Sbjct: 164 HHLGYHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHH 199
Score = 51.2 bits (121), Expect = 8e-06
Identities = 18/44 (40%), Positives = 20/44 (44%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
H P +H H HHH + HHHHH HH H HH H
Sbjct: 209 HHHHAPHHHHHHHHGHHHHHHHHHGHHHHHHHHHGHHHHHHHHH 252
Score = 51.2 bits (121), Expect = 8e-06
Identities = 17/33 (51%), Positives = 20/33 (60%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
+H + H HHH + HHH HH HH HH HH H
Sbjct: 217 HHHHHHGHHHHHHHHHGHHHHHHHHHGHHHHHH 249
Score = 51.2 bits (121), Expect = 8e-06
Identities = 18/36 (50%), Positives = 20/36 (55%), Gaps = 4/36 (11%)
Frame = -3
Query: 325 HWNFHFHHHSYFHHHHH----HRHHLHHAQPHHRRH 230
H + H HHH + HHHHH H HH HH HH H
Sbjct: 234 HHHHHHHHHGHHHHHHHHHDAHHHHHHHHDAHHHHH 269
Score = 50.8 bits (120), Expect = 1e-05
Identities = 20/53 (37%), Positives = 26/53 (48%), Gaps = 9/53 (16%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYFHHHH---------HHRHHLHHAQPHHRRH 230
H+ ++ +H + HHH +FHHHH HH HH HHA HH H
Sbjct: 138 HAAHHHHHEEHHHHHHAAHHHPWFHHHHLGYHHHHAPHHHHHHHHAPHHHHHH 190
Score = 50.8 bits (120), Expect = 1e-05
Identities = 17/33 (51%), Positives = 20/33 (60%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
+H + H HHH + H HHHH HH H A HH H
Sbjct: 229 HHHHGHHHHHHHHHGHHHHHHHHHDAHHHHHHH 261
Score = 50.4 bits (119), Expect = 1e-05
Identities = 21/54 (38%), Positives = 25/54 (45%), Gaps = 7/54 (12%)
Frame = -3
Query: 370 ISIHSERDPKSQKMNHW---NFHFHHHSYFHHHHHH----RHHLHHAQPHHRRH 230
+ H P +H + H HHH+ HHHHHH HH HH PHH H
Sbjct: 166 LGYHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHH 219
Score = 50.1 bits (118), Expect = 2e-05
Identities = 19/46 (41%), Positives = 22/46 (47%), Gaps = 2/46 (4%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYF--HHHHHHRHHLHHAQPHHRRH 230
H D +H + H HHH + HHHHHH H HH HH H
Sbjct: 269 HHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHDAH 314
Score = 49.7 bits (117), Expect = 2e-05
Identities = 18/36 (50%), Positives = 19/36 (52%), Gaps = 4/36 (11%)
Frame = -3
Query: 325 HWNFHFHHHSYFHHHHH----HRHHLHHAQPHHRRH 230
H + H HHH HHHHH H HH HH HH H
Sbjct: 244 HHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHH 279
Score = 49.7 bits (117), Expect = 2e-05
Identities = 18/35 (51%), Positives = 21/35 (59%), Gaps = 2/35 (5%)
Frame = -3
Query: 328 NHWNFHFHHHSYFHHH--HHHRHHLHHAQPHHRRH 230
+H + H HHH + HHH HHH HH H A HH H
Sbjct: 237 HHHHHHGHHHHHHHHHDAHHHHHHHHDAHHHHHHH 271
Score = 49.3 bits (116), Expect = 3e-05
Identities = 18/44 (40%), Positives = 20/44 (44%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
H P +H H HHH + HHHHH HH H HH H
Sbjct: 199 HHHHAPHHHHHHHHAPHHHHHHHHGHHHHHHHHHGHHHHHHHHH 242
Score = 48.5 bits (114), Expect = 5e-05
Identities = 17/35 (48%), Positives = 20/35 (56%), Gaps = 3/35 (8%)
Frame = -3
Query: 325 HWNFHFHHHSYFHHHHHH---RHHLHHAQPHHRRH 230
H + H HH ++ HHHHHH HH HH HH H
Sbjct: 314 HHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHH 348
Score = 48.1 bits (113), Expect = 7e-05
Identities = 18/43 (41%), Positives = 21/43 (47%), Gaps = 2/43 (4%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYF--HHHHHHRHHLHHAQPHH 239
H D +H + H HHH + HHHHHH H HH HH
Sbjct: 249 HHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHH 291
Score = 48.1 bits (113), Expect = 7e-05
Identities = 18/43 (41%), Positives = 21/43 (47%), Gaps = 2/43 (4%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYF--HHHHHHRHHLHHAQPHH 239
H D +H + H HHH + HHHHHH H HH HH
Sbjct: 308 HHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHH 350
Score = 48.1 bits (113), Expect = 7e-05
Identities = 19/46 (41%), Positives = 22/46 (47%), Gaps = 2/46 (4%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYFHHHHHHRHH--LHHAQPHHRRH 230
H D +H + H HHH + HHHHH HH HH HH H
Sbjct: 289 HHHHDAHHHHHHHHDAHHHHHHHDAHHHHHHHHDAHHHHHHHHDAH 334
Score = 47.8 bits (112), Expect = 9e-05
Identities = 20/52 (38%), Positives = 24/52 (45%), Gaps = 8/52 (15%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHH----SYFHHHHHH----RHHLHHAQPHHRRH 230
H D +H + H HHH ++ HHHHHH HH HH HH H
Sbjct: 259 HHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHH 310
Score = 47.4 bits (111), Expect = 1e-04
Identities = 17/36 (47%), Positives = 20/36 (55%), Gaps = 4/36 (11%)
Frame = -3
Query: 325 HWNFHFHHHSYFHHHHHH----RHHLHHAQPHHRRH 230
H + H HH ++ HHHHHH HH HH HH H
Sbjct: 255 HHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHH 290
Score = 47.4 bits (111), Expect = 1e-04
Identities = 19/46 (41%), Positives = 21/46 (45%), Gaps = 2/46 (4%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYF--HHHHHHRHHLHHAQPHHRRH 230
H P +H H HHH + HHHHHH H HH HH H
Sbjct: 179 HHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHGH 224
Score = 47.0 bits (110), Expect = 2e-04
Identities = 20/52 (38%), Positives = 24/52 (45%), Gaps = 8/52 (15%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHH----SYFHHHHH----HRHHLHHAQPHHRRH 230
H D +H + H HHH ++ HHHHH H HH H A HH H
Sbjct: 279 HHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHDAHHHHHHHHDAHHHHHHH 330
Score = 46.6 bits (109), Expect = 2e-04
Identities = 19/46 (41%), Positives = 21/46 (45%), Gaps = 2/46 (4%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYFHHHHHHRHH--LHHAQPHHRRH 230
H D +H H HHH + HHHHH HH HH HH H
Sbjct: 299 HHHHDAHHHHHHHDAHHHHHHHHDAHHHHHHHHDAHHHHHHHHDAH 344
Score = 45.4 bits (106), Expect = 5e-04
Identities = 17/44 (38%), Positives = 19/44 (42%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYFHHHHHHRHHLHHAQPHHRRH 230
H P +H H HHH + HHHH HH H HH H
Sbjct: 189 HHHHAPHHHHHHHHAPHHHHHHHHAPHHHHHHHHGHHHHHHHHH 232
Score = 42.4 bits (98), Expect = 0.004
Identities = 19/50 (38%), Positives = 22/50 (44%), Gaps = 6/50 (12%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYFHHHHH---HRHHL---HHAQPHHRRH 230
H E +H + HHH + HHH H HHL HH PHH H
Sbjct: 130 HEEHHHHHHAAHHHHHEEHHHHHHAAHHHPWFHHHHLGYHHHHAPHHHHH 179
Score = 41.6 bits (96), Expect = 0.007
Identities = 20/61 (32%), Positives = 22/61 (35%), Gaps = 17/61 (27%)
Frame = -3
Query: 361 HSERDPKSQKMNHWNFHFHHHSYFH--------HHHH---------HRHHLHHAQPHHRR 233
H E H + H HH + H HHHH H HH HH PHH
Sbjct: 129 HHEEHHHHHHAAHHHHHEEHHHHHHAAHHHPWFHHHHLGYHHHHAPHHHHHHHHAPHHHH 188
Query: 232 H 230
H
Sbjct: 189 H 189
Score = 38.9 bits (89), Expect = 0.043
Identities = 19/55 (34%), Positives = 24/55 (43%), Gaps = 9/55 (16%)
Frame = -3
Query: 367 SIHSERDPKSQKMNHWNFHF--HHHSYFHHH---HHHR----HHLHHAQPHHRRH 230
++H E + +H H HH + HHH HH HH HH PHH H
Sbjct: 51 TVHPEHLHEEHHHHHPEEHHEPHHEEHHHHHPEEHHEPHHEEHHHHHPHPHHHHH 105
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 590,250,967
Number of Sequences: 1393205
Number of extensions: 17998362
Number of successful extensions: 780144
Number of sequences better than 10.0: 12032
Number of HSP's better than 10.0 without gapping: 124248
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 404044
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)