Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011947A_C01 KMC011947A_c01
(907 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
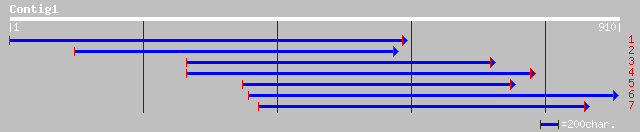
gb|AAD54273.1|AF167556_1 dihydroflavonol-4-reductase DFR1 [Glyci... 244 2e-63
gb|AAN63056.1| dihydroflavonol reductase [Populus tremuloides] 217 2e-55
emb|CAB94914.1| dihydroflavonol 4-reductase [Juglans nigra] 216 3e-55
emb|CAA72420.1| dihydroflavonol 4-reductase [Vitis vinifera] 214 1e-54
gb|AAM73809.1|AF449422_1 dihydroflavonol-4-reductase [Solanum tu... 213 4e-54
>gb|AAD54273.1|AF167556_1 dihydroflavonol-4-reductase DFR1 [Glycine max]
Length = 347
Score = 244 bits (622), Expect = 2e-63
Identities = 111/138 (80%), Positives = 131/138 (94%)
Frame = -3
Query: 905 VVGPFLMPTMPPSLITALSLITGNEAHYSIIKQGQYVHLDDLCLAHIFLFENPKAQGRYM 726
VVGPFLMPTMPPSLITALS ITGNE HYSIIKQGQ+VHLDDLCLAHIFLFE P+ +GRY+
Sbjct: 193 VVGPFLMPTMPPSLITALSPITGNEDHYSIIKQGQFVHLDDLCLAHIFLFEEPEVEGRYI 252
Query: 725 CSAYEATIHEVARMINKKYPEFNVPTKFKDIPDELDIIKFSSKKITDLGFKFKYSLEDMY 546
CSA +ATIH++A++IN+KYPE+ VPTKFK+IPD+L++++FSSKKITDLGFKFKYSLEDMY
Sbjct: 253 CSACDATIHDIAKLINQKYPEYKVPTKFKNIPDQLELVRFSSKKITDLGFKFKYSLEDMY 312
Query: 545 TGAVETCREKGLLPKTAE 492
TGA++TCR+KGLLPK AE
Sbjct: 313 TGAIDTCRDKGLLPKPAE 330
>gb|AAN63056.1| dihydroflavonol reductase [Populus tremuloides]
Length = 346
Score = 217 bits (553), Expect = 2e-55
Identities = 96/134 (71%), Positives = 121/134 (89%)
Frame = -3
Query: 905 VVGPFLMPTMPPSLITALSLITGNEAHYSIIKQGQYVHLDDLCLAHIFLFENPKAQGRYM 726
VVGPF+M +MPPSL+TALSLITGNEAHY I+KQG YVHLDDLC++HIFL+ENPKA+GRY+
Sbjct: 193 VVGPFIMQSMPPSLLTALSLITGNEAHYGILKQGHYVHLDDLCMSHIFLYENPKAEGRYI 252
Query: 725 CSAYEATIHEVARMINKKYPEFNVPTKFKDIPDELDIIKFSSKKITDLGFKFKYSLEDMY 546
C++ +A IH++A+++ +KYPE+NVP KFKDI + L + FSSKK+TDLGF+FKYSLEDM+
Sbjct: 253 CNSDDANIHDLAQLLREKYPEYNVPDKFKDIDENLACVAFSSKKLTDLGFEFKYSLEDMF 312
Query: 545 TGAVETCREKGLLP 504
GAVETCREKGL+P
Sbjct: 313 AGAVETCREKGLIP 326
>emb|CAB94914.1| dihydroflavonol 4-reductase [Juglans nigra]
Length = 229
Score = 216 bits (551), Expect = 3e-55
Identities = 101/138 (73%), Positives = 122/138 (88%)
Frame = -3
Query: 905 VVGPFLMPTMPPSLITALSLITGNEAHYSIIKQGQYVHLDDLCLAHIFLFENPKAQGRYM 726
VVGPFLMP+MPPSLITALS ITGNEAHYSIIKQGQ+VHLDDLC+ HI+LFE+PKA+GRY+
Sbjct: 89 VVGPFLMPSMPPSLITALSPITGNEAHYSIIKQGQFVHLDDLCMGHIYLFEHPKAEGRYL 148
Query: 725 CSAYEATIHEVARMINKKYPEFNVPTKFKDIPDELDIIKFSSKKITDLGFKFKYSLEDMY 546
CSA +ATI +VA+++ +K+PE NVPTKFK + + L+II F+SKKI DLGF+FKYSLEDM+
Sbjct: 149 CSACDATILDVAKLLREKFPECNVPTKFKGVDESLEIISFNSKKIKDLGFQFKYSLEDMF 208
Query: 545 TGAVETCREKGLLPKTAE 492
AV+TCR KGLLP AE
Sbjct: 209 VEAVQTCRAKGLLPPAAE 226
>emb|CAA72420.1| dihydroflavonol 4-reductase [Vitis vinifera]
Length = 337
Score = 214 bits (546), Expect = 1e-54
Identities = 98/145 (67%), Positives = 124/145 (84%)
Frame = -3
Query: 905 VVGPFLMPTMPPSLITALSLITGNEAHYSIIKQGQYVHLDDLCLAHIFLFENPKAQGRYM 726
VVGPF+M +MPPSLITALS ITGNEAHYSII+QGQ+VHLDDLC AHI+LFENPKA+GRY+
Sbjct: 193 VVGPFIMSSMPPSLITALSPITGNEAHYSIIRQGQFVHLDDLCNAHIYLFENPKAEGRYI 252
Query: 725 CSAYEATIHEVARMINKKYPEFNVPTKFKDIPDELDIIKFSSKKITDLGFKFKYSLEDMY 546
CS+++ I ++A+M+ +KYPE+N+PT+FK + + L + FSSKK+TDLGF+FKYSLEDM+
Sbjct: 253 CSSHDCIILDLAKMLREKYPEYNIPTEFKGVDENLKSVCFSSKKLTDLGFEFKYSLEDMF 312
Query: 545 TGAVETCREKGLLPKTAETPATNGT 471
TGAV+TCR KGLLP + E P T
Sbjct: 313 TGAVDTCRAKGLLPPSHEKPVDGKT 337
>gb|AAM73809.1|AF449422_1 dihydroflavonol-4-reductase [Solanum tuberosum]
Length = 382
Score = 213 bits (541), Expect = 4e-54
Identities = 98/155 (63%), Positives = 127/155 (81%)
Frame = -3
Query: 905 VVGPFLMPTMPPSLITALSLITGNEAHYSIIKQGQYVHLDDLCLAHIFLFENPKAQGRYM 726
VVGPF+ PT PPSLITALSLITGNEAHY IIKQGQYVHLDDLC AHIFL+E+PKA+GR++
Sbjct: 205 VVGPFITPTFPPSLITALSLITGNEAHYGIIKQGQYVHLDDLCEAHIFLYEHPKAEGRFI 264
Query: 725 CSAYEATIHEVARMINKKYPEFNVPTKFKDIPDELDIIKFSSKKITDLGFKFKYSLEDMY 546
CS++ A I++VA+M+ +K+PE+ VPT+FK I +L I+ FSSKK+ D+GF+FKY+LEDMY
Sbjct: 265 CSSHHAIIYDVAKMVRQKWPEYYVPTEFKGIDKDLPIVSFSSKKLMDMGFQFKYTLEDMY 324
Query: 545 TGAVETCREKGLLPKTAETPATNGTTQK*MCFLTQ 441
GA+ETCR+K LLP + A NG ++ + T+
Sbjct: 325 KGAIETCRQKQLLPFHTRSTAANGKDKEAIPIFTE 359
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 745,081,994
Number of Sequences: 1393205
Number of extensions: 15700646
Number of successful extensions: 35632
Number of sequences better than 10.0: 205
Number of HSP's better than 10.0 without gapping: 34277
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35510
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 49363855696
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)