Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011917A_C01 KMC011917A_c01
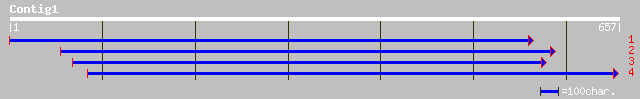
(657 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC42257.1| putative carboxyphosphonoenolpyruvate mutase [Ar... 241 6e-63
ref|NP_181847.1| putative carboxyphosphonoenolpyruvate mutase; p... 229 2e-59
ref|ZP_00074372.1| hypothetical protein [Trichodesmium erythraeu... 173 2e-42
ref|NP_485903.1| carboxyphosphonoenolpyruvate phosphonomutase [N... 162 4e-39
ref|ZP_00030593.1| hypothetical protein [Burkholderia fungorum] 155 6e-37
>dbj|BAC42257.1| putative carboxyphosphonoenolpyruvate mutase [Arabidopsis thaliana]
gi|28827730|gb|AAO50709.1| putative
carboxyphosphonoenolpyruvate mutase [Arabidopsis
thaliana]
Length = 479
Score = 241 bits (615), Expect = 6e-63
Identities = 121/219 (55%), Positives = 169/219 (76%), Gaps = 2/219 (0%)
Frame = +1
Query: 4 YSSSISICRRSPAKTHTSLHTSGSSSRKMEKG-VKALRRLLQSPGVHQGPIAFNALSAKL 180
+++ S+ RR ++++ + S + K+ K LR ++QSPGV QGP F+ALSAKL
Sbjct: 40 FANPFSVSRRFRGAVNSTVVSDESGNAKLPSSPAKKLRNIMQSPGVLQGPCCFDALSAKL 99
Query: 181 VESTGFPFIFTGGFALSATRLALPDAGLISYGEMVDQGRLVTQAISVPVIGDADTGFGNS 360
+E GFP+ T GF++SA+RL LPD GLISYGEMVDQG+ +TQ++S+PVIGD G+GN+
Sbjct: 100 IERAGFPYCITSGFSISASRLGLPDKGLISYGEMVDQGQQITQSVSIPVIGDGGNGYGNA 159
Query: 361 VNVKRTVKGFIDAGFAGIMLEDQISPKACGHTQG-RKVVSREEALMRIKAAVDARRESGS 537
+NVKRTVKG+I AGFAGI++ D++ C +T+ R+VVSREEA+MR+KAAVDARRE S
Sbjct: 160 MNVKRTVKGYIKAGFAGIIINDEV---CCENTKSERRVVSREEAVMRVKAAVDARRECDS 216
Query: 538 DIVIVARSDARQAVSLDEAFIRTKAFAEIGADVLFIDAL 654
DIVIVA++D+R+A+SL+E+ IR +AF + GADVL +D+L
Sbjct: 217 DIVIVAQTDSREAISLEESLIRARAFTDAGADVLSVDSL 255
>ref|NP_181847.1| putative carboxyphosphonoenolpyruvate mutase; protein id:
At2g43180.1 [Arabidopsis thaliana]
gi|25408843|pir||A84863 probable
carboxyphosphonoenolpyruvate mutase [imported] -
Arabidopsis thaliana gi|3763927|gb|AAC64307.1| putative
carboxyphosphonoenolpyruvate mutase [Arabidopsis
thaliana]
Length = 492
Score = 229 bits (584), Expect = 2e-59
Identities = 119/219 (54%), Positives = 165/219 (75%), Gaps = 2/219 (0%)
Frame = +1
Query: 4 YSSSISICRRSPAKTHTSLHTSGSSSRKMEKG-VKALRRLLQSPGVHQGPIAFNALSAKL 180
+++ S+ RR ++++ + S + K+ K LR ++QSPGV QGP F+ALSAKL
Sbjct: 40 FANPFSVSRRFRGAVNSTVVSDESGNAKLPSSPAKKLRNIMQSPGVLQGPCCFDALSAKL 99
Query: 181 VESTGFPFIFTGGFALSATRLALPDAGLISYGEMVDQGRLVTQAISVPVIGDADTGFGNS 360
+E GFP+ T SA+RL LPD GLISYGEMVDQG+ +TQ++S+PVIGD G+GN+
Sbjct: 100 IERAGFPYCIT-----SASRLGLPDKGLISYGEMVDQGQQITQSVSIPVIGDGGNGYGNA 154
Query: 361 VNVKRTVKGFIDAGFAGIMLEDQISPKACGHTQG-RKVVSREEALMRIKAAVDARRESGS 537
+NVKRTVKG+I AGFAGI++ D++ C +T+ R+VVSREEA+MR+KAAVDARRE S
Sbjct: 155 MNVKRTVKGYIKAGFAGIIINDEV---CCENTKSERRVVSREEAVMRVKAAVDARRECDS 211
Query: 538 DIVIVARSDARQAVSLDEAFIRTKAFAEIGADVLFIDAL 654
DIVIVA++D+R+A+SL+E+ IR +AF + GADVL +D+L
Sbjct: 212 DIVIVAQTDSREAISLEESLIRARAFTDAGADVLSVDSL 250
>ref|ZP_00074372.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 291
Score = 173 bits (438), Expect = 2e-42
Identities = 88/183 (48%), Positives = 126/183 (68%)
Frame = +1
Query: 103 KALRRLLQSPGVHQGPIAFNALSAKLVESTGFPFIFTGGFALSATRLALPDAGLISYGEM 282
K LR++L+ PG P ++ + AK+VE GFP +FT GF +S + L PD G I+ EM
Sbjct: 5 KKLRQILEQPGALVLPGVYDCIGAKIVEQIGFPVVFTSGFGISGSTLGRPDYGFITATEM 64
Query: 283 VDQGRLVTQAISVPVIGDADTGFGNSVNVKRTVKGFIDAGFAGIMLEDQISPKACGHTQG 462
+ R +T+++++P++ D DTG+GN +NV RTV ++ G AGI+LEDQ PK CGH QG
Sbjct: 65 LYAIRRITESVNIPLVADIDTGYGNPLNVIRTVTDIVNMGVAGIILEDQEWPKKCGHFQG 124
Query: 463 RKVVSREEALMRIKAAVDARRESGSDIVIVARSDARQAVSLDEAFIRTKAFAEIGADVLF 642
++V+ E + +IKAAV AR +SG +VI+AR+DAR + LDEA R +A AE GADV+F
Sbjct: 125 KRVIPMAEHVEKIKAAVHARGDSG--LVIIARTDARAPLGLDEAIKRGRACAEAGADVVF 182
Query: 643 IDA 651
I+A
Sbjct: 183 IEA 185
>ref|NP_485903.1| carboxyphosphonoenolpyruvate phosphonomutase [Nostoc sp. PCC 7120]
gi|25324632|pir||AI2038 carboxyphosphonoenolpyruvate
phosphonomutase [imported] - Nostoc sp. (strain PCC
7120) gi|17130953|dbj|BAB73562.1|
carboxyphosphonoenolpyruvate phosphonomutase [Nostoc sp.
PCC 7120]
Length = 287
Score = 162 bits (410), Expect = 4e-39
Identities = 82/181 (45%), Positives = 123/181 (67%)
Frame = +1
Query: 109 LRRLLQSPGVHQGPIAFNALSAKLVESTGFPFIFTGGFALSATRLALPDAGLISYGEMVD 288
LR+LL +P + P ++ LSAKL E+ GF + T GF ++A+ L LPD G ++ E +
Sbjct: 7 LRQLLANPEIIVIPGIYDCLSAKLAENIGFDVVATSGFGIAASTLGLPDYGFLTATEALY 66
Query: 289 QGRLVTQAISVPVIGDADTGFGNSVNVKRTVKGFIDAGFAGIMLEDQISPKACGHTQGRK 468
+ Q++SVP+I D DTG+GN++NV RT+K + G AG++LEDQ PK CGH +G++
Sbjct: 67 SVGRIAQSVSVPLIADLDTGYGNALNVMRTIKDAVQLGVAGVLLEDQEWPKKCGHFEGKR 126
Query: 469 VVSREEALMRIKAAVDARRESGSDIVIVARSDARQAVSLDEAFIRTKAFAEIGADVLFID 648
V+ E +I+AAV+AR +SG +VI+AR+DAR + L+EA R A+ E GAD+LF++
Sbjct: 127 VIPTSEHAGKIRAAVEARGDSG--LVIIARTDARGPLGLEEAIARGHAYIEAGADILFVE 184
Query: 649 A 651
A
Sbjct: 185 A 185
>ref|ZP_00030593.1| hypothetical protein [Burkholderia fungorum]
Length = 296
Score = 155 bits (391), Expect = 6e-37
Identities = 76/181 (41%), Positives = 118/181 (64%)
Frame = +1
Query: 109 LRRLLQSPGVHQGPIAFNALSAKLVESTGFPFIFTGGFALSATRLALPDAGLISYGEMVD 288
L+ +L+S P F+ SA++ + F ++ G+ +S + L + DAGL+SY +MV
Sbjct: 7 LKAILKSGRFVVAPGVFDMFSARVADRLPFEALYMTGYGVSGSYLGVADAGLVSYADMVG 66
Query: 289 QGRLVTQAISVPVIGDADTGFGNSVNVKRTVKGFIDAGFAGIMLEDQISPKACGHTQGRK 468
+ R + + P+I DADTGFG +NV+ TV+G+ AG I +EDQ+ PK CGHTQGR+
Sbjct: 67 RARQIAEGTGKPLIADADTGFGGLLNVRHTVRGYEAAGVQAIQMEDQVMPKKCGHTQGRQ 126
Query: 469 VVSREEALMRIKAAVDARRESGSDIVIVARSDARQAVSLDEAFIRTKAFAEIGADVLFID 648
VV + L +I+ A++ARR D++I+AR+D+R + LDEA R KAFA GAD++F++
Sbjct: 127 VVPLNDMLKKIEVALEARR--SDDMLIIARTDSRTTLGLDEAIRRGKAFARAGADIVFVE 184
Query: 649 A 651
+
Sbjct: 185 S 185
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 603,948,029
Number of Sequences: 1393205
Number of extensions: 14857588
Number of successful extensions: 87818
Number of sequences better than 10.0: 542
Number of HSP's better than 10.0 without gapping: 69972
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 83770
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28006887348
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)