Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011915A_C01 KMC011915A_c01
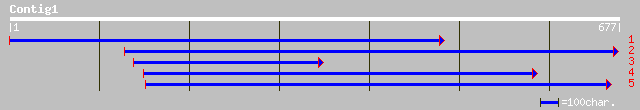
(676 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T00093 hypothetical protein KIAA0467 - human (fragment) gi|... 33 3.6
gb|AAD15618.1| similar to phosphatidylinositol (4,5)bisphosphate... 33 4.6
ref|NP_649341.1| CG7162-PA [Drosophila melanogaster] gi|9937476|... 33 4.6
gb|AAM66074.1| endosperm-specific protein-like protein [Arabidop... 32 6.1
ref|NP_566043.1| fasciclin-like arabinogalactan-protein (FLA8); ... 32 6.1
>pir||T00093 hypothetical protein KIAA0467 - human (fragment)
gi|3413896|dbj|BAA32312.1| KIAA0467 protein [Homo
sapiens]
Length = 2055
Score = 33.1 bits (74), Expect = 3.6
Identities = 17/42 (40%), Positives = 25/42 (59%)
Frame = -1
Query: 526 ELSLVQSSITCPAQSPSGSLDFGAVNEYSGLGKAKDKPPGPV 401
EL L+ +S+ C +P+GSL G++ S G+A PP PV
Sbjct: 970 ELQLLPASL-CTEDTPTGSLRNGSLETKSSAGRASTFPPAPV 1010
>gb|AAD15618.1| similar to phosphatidylinositol (4,5)bisphosphate 5-phosphatase;
match to PID:g1399105 [Homo sapiens]
Length = 1056
Score = 32.7 bits (73), Expect = 4.6
Identities = 26/81 (32%), Positives = 36/81 (44%), Gaps = 1/81 (1%)
Frame = -1
Query: 676 PISQPNLGAPNIAPSASGKGNRKLSSDG-KPGSFDSGCALYLLSTLQSQSPELSLVQSSI 500
P S P PN+AP++ + +S G KP SG +L L S + Q PEL S +
Sbjct: 160 PRSPPVTLGPNLAPTSRDQKQEPPASVGPKPTLAASGLSLALAS--EEQPPELPSTPSPV 217
Query: 499 TCPAQSPSGSLDFGAVNEYSG 437
P SP+ + SG
Sbjct: 218 PSPVLSPTQEQALAPASTASG 238
>ref|NP_649341.1| CG7162-PA [Drosophila melanogaster]
gi|9937476|gb|AAG02461.1|AF289996_1 thyroid hormone
receptor-associated protein TRAP220 [Drosophila
melanogaster] gi|23094268|gb|AAF51757.2| CG7162-PA
[Drosophila melanogaster]
Length = 1475
Score = 32.7 bits (73), Expect = 4.6
Identities = 20/50 (40%), Positives = 23/50 (46%)
Frame = -1
Query: 676 PISQPNLGAPNIAPSASGKGNRKLSSDGKPGSFDSGCALYLLSTLQSQSP 527
P +P++GA S G G RKL S GS SG LS SQ P
Sbjct: 1300 PRRRPSMGALASGSSGGGSGQRKLGSASGGGSGSSGSVSPALSGSMSQPP 1349
>gb|AAM66074.1| endosperm-specific protein-like protein [Arabidopsis thaliana]
Length = 420
Score = 32.3 bits (72), Expect = 6.1
Identities = 18/41 (43%), Positives = 23/41 (55%)
Frame = +3
Query: 183 PESFNKSKLNYWSTRFYFEDQQLHKFTKDTTPLTTYSQTKG 305
P S KS L+ Y++ Q+LHK +K TT TT QT G
Sbjct: 78 PLSVIKSALSLLVLLDYYDPQKLHKISKGTTLSTTLYQTTG 118
>ref|NP_566043.1| fasciclin-like arabinogalactan-protein (FLA8); protein id:
At2g45470.1, supported by cDNA: 7709., supported by
cDNA: gi_20453146 [Arabidopsis thaliana]
gi|25349350|pir||H84890 hypothetical protein At2g45470
[imported] - Arabidopsis thaliana
gi|2583108|gb|AAB82617.1| expressed protein [Arabidopsis
thaliana] gi|20453147|gb|AAM19815.1| At2g45470/F4L23.2
[Arabidopsis thaliana] gi|23506197|gb|AAN31110.1|
At2g45470/F4L23.2 [Arabidopsis thaliana]
Length = 420
Score = 32.3 bits (72), Expect = 6.1
Identities = 18/41 (43%), Positives = 23/41 (55%)
Frame = +3
Query: 183 PESFNKSKLNYWSTRFYFEDQQLHKFTKDTTPLTTYSQTKG 305
P S KS L+ Y++ Q+LHK +K TT TT QT G
Sbjct: 78 PLSVIKSALSLLVLLDYYDPQKLHKISKGTTLSTTLYQTTG 118
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 594,639,991
Number of Sequences: 1393205
Number of extensions: 13399385
Number of successful extensions: 37548
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 36011
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37530
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29704274460
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)