Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
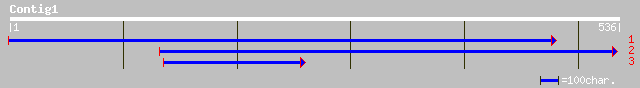
Query= KMC011840A_C01 KMC011840A_c01
(535 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_565588.1| expressed protein; protein id: At2g25190.1, sup... 38 0.066
ref|NP_194926.1| putative protein; protein id: At4g31980.1 [Arab... 38 0.066
ref|NP_568467.1| putative protein; protein id: At5g25170.1, supp... 38 0.066
gb|AAL38722.1| unknown protein [Arabidopsis thaliana] gi|2025907... 38 0.066
ref|NP_720427.1| conserved hypothetical protein [Shewanella onei... 33 2.8
>ref|NP_565588.1| expressed protein; protein id: At2g25190.1, supported by cDNA:
14105., supported by cDNA: gi_13877550, supported by
cDNA: gi_20148724 [Arabidopsis thaliana]
gi|25354456|pir||D84645 hypothetical protein At2g25190
[imported] - Arabidopsis thaliana
gi|4567258|gb|AAD23672.1| expressed protein [Arabidopsis
thaliana] gi|13877551|gb|AAK43853.1|AF370476_1 Unknown
protein [Arabidopsis thaliana]
gi|20148725|gb|AAM10253.1| unknown protein [Arabidopsis
thaliana] gi|21553378|gb|AAM62471.1| unknown
[Arabidopsis thaliana]
Length = 240
Score = 38.1 bits (87), Expect = 0.066
Identities = 19/40 (47%), Positives = 26/40 (64%), Gaps = 3/40 (7%)
Frame = -1
Query: 535 LGFLCNCVLPPSLNE---KKVGPAALTHRIPEEGKNKMRS 425
LG LCNCVLPP LNE ++VG L+ ++ +N+ RS
Sbjct: 142 LGVLCNCVLPPRLNEAKVRRVGKGELSESEKKKLRNRSRS 181
>ref|NP_194926.1| putative protein; protein id: At4g31980.1 [Arabidopsis thaliana]
gi|7485367|pir||T04647 hypothetical protein F10N7.210 -
Arabidopsis thaliana gi|2827637|emb|CAA16591.1| putative
protein [Arabidopsis thaliana]
gi|7270102|emb|CAB79916.1| putative protein [Arabidopsis
thaliana]
Length = 680
Score = 38.1 bits (87), Expect = 0.066
Identities = 18/33 (54%), Positives = 21/33 (63%)
Frame = -1
Query: 535 LGFLCNCVLPPSLNEKKVGPAALTHRIPEEGKN 437
LGFLCNCVLP LNE KV ++ EG+N
Sbjct: 142 LGFLCNCVLPACLNETKVKRVGKDGKLLLEGEN 174
>ref|NP_568467.1| putative protein; protein id: At5g25170.1, supported by cDNA:
263500. [Arabidopsis thaliana]
Length = 218
Score = 38.1 bits (87), Expect = 0.066
Identities = 20/39 (51%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Frame = -1
Query: 532 GFLCNCVLPPSLNEKKVGPA-ALTHRIPEEGKNKMRSPS 419
G CNCVLP LNE KV + +IPE K K+RS S
Sbjct: 144 GLFCNCVLPAELNETKVRQVRSKEEKIPEVEKKKLRSRS 182
>gb|AAL38722.1| unknown protein [Arabidopsis thaliana] gi|20259079|gb|AAM14255.1|
unknown protein [Arabidopsis thaliana]
Length = 249
Score = 38.1 bits (87), Expect = 0.066
Identities = 18/33 (54%), Positives = 21/33 (63%)
Frame = -1
Query: 535 LGFLCNCVLPPSLNEKKVGPAALTHRIPEEGKN 437
LGFLCNCVLP LNE KV ++ EG+N
Sbjct: 142 LGFLCNCVLPACLNETKVKRVGKDGKLLLEGEN 174
>ref|NP_720427.1| conserved hypothetical protein [Shewanella oneidensis MR-1]
gi|24345216|gb|AAN53027.1|AE015916_2 conserved
hypothetical protein [Shewanella oneidensis MR-1]
Length = 577
Score = 32.7 bits (73), Expect = 2.8
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Frame = +3
Query: 393 DWRKPHAWWDGL--LILFFPSSGILWVNAAGPTFFSFR 500
DW K H W DGL ++L F SS +L NAA P + R
Sbjct: 211 DWSKNHFWEDGLFSVVLPFMSSFLLDPNAADPGYLKTR 248
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 460,216,188
Number of Sequences: 1393205
Number of extensions: 9967488
Number of successful extensions: 26087
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 25076
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26065
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17885181664
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)