Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011824A_C01 KMC011824A_c01
(629 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
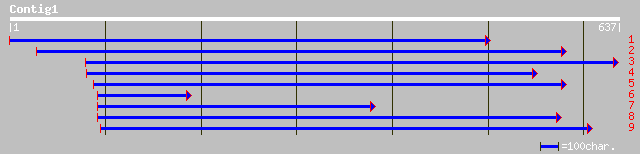
emb|CAA09854.1| cyclin D3.2 protein [Nicotiana tabacum] 80 3e-14
gb|AAO13248.1|AF181993_1 cyclin D [Populus tremula x Populus tre... 79 6e-14
gb|AAM77273.1|AF519810_1 cyclin D3.1 protein [Lagenaria siceraria] 78 8e-14
emb|CAB60838.1| CycD3;3 [Lycopersicon esculentum] 76 4e-13
emb|CAB61223.1| cyclin D3b [Antirrhinum majus] 75 7e-13
>emb|CAA09854.1| cyclin D3.2 protein [Nicotiana tabacum]
Length = 367
Score = 79.7 bits (195), Expect = 3e-14
Identities = 41/78 (52%), Positives = 57/78 (72%), Gaps = 2/78 (2%)
Frame = -1
Query: 626 DECCKVMLEMWSGIDDQGNKLC--MKRKFGLIPGSPNGVVDVCFSCDSSNDSWAVAVNSV 453
+EC ++LE+ + G +C +KRK +PGSP+GV+D FSCDSSNDSW+VA +S+
Sbjct: 277 EECHDLILEL---MGTSGYNICQSLKRKHQSVPGSPSGVIDAYFSCDSSNDSWSVA-SSI 332
Query: 452 TSSPEPLPKKIRTQDQLM 399
+SSPEP K+I+TQDQ M
Sbjct: 333 SSSPEPQYKRIKTQDQTM 350
>gb|AAO13248.1|AF181993_1 cyclin D [Populus tremula x Populus tremuloides]
Length = 376
Score = 78.6 bits (192), Expect = 6e-14
Identities = 47/90 (52%), Positives = 60/90 (66%)
Frame = -1
Query: 629 VDECCKVMLEMWSGIDDQGNKLCMKRKFGLIPGSPNGVVDVCFSCDSSNDSWAVAVNSVT 450
V++C K ++E + D L KRKF +PGSP+GVVDV FS DSSNDSW+VA +SV+
Sbjct: 288 VEDCSKFLMEF--ALRDHFKLLSNKRKFCSLPGSPSGVVDVSFSSDSSNDSWSVA-SSVS 344
Query: 449 SSPEPLPKKIRTQDQLMQNHFHL*FSKHSS 360
SSP+PL KK R Q + N FS+HSS
Sbjct: 345 SSPKPLSKKSRAL-QSLNNATTSDFSQHSS 373
>gb|AAM77273.1|AF519810_1 cyclin D3.1 protein [Lagenaria siceraria]
Length = 352
Score = 78.2 bits (191), Expect = 8e-14
Identities = 42/76 (55%), Positives = 51/76 (66%)
Frame = -1
Query: 629 VDECCKVMLEMWSGIDDQGNKLCMKRKFGLIPGSPNGVVDVCFSCDSSNDSWAVAVNSVT 450
V+ECCK++ S + KRK G IPGSPNGV+DV FS DSSNDSW+VA +SV+
Sbjct: 271 VEECCKLI----SNASRRNGNQFKKRKIGSIPGSPNGVMDVSFSSDSSNDSWSVA-SSVS 325
Query: 449 SSPEPLPKKIRTQDQL 402
SSPEPL KK R +
Sbjct: 326 SSPEPLTKKNRANGSM 341
>emb|CAB60838.1| CycD3;3 [Lycopersicon esculentum]
Length = 336
Score = 75.9 bits (185), Expect = 4e-13
Identities = 41/75 (54%), Positives = 52/75 (68%)
Frame = -1
Query: 629 VDECCKVMLEMWSGIDDQGNKLCMKRKFGLIPGSPNGVVDVCFSCDSSNDSWAVAVNSVT 450
V+ C +++ E+ ID NK RKFG +PGSP GV+D+ FS D SNDSW+VA SVT
Sbjct: 267 VEGCYRLIQEVACNIDFGSNK----RKFGTLPGSPTGVMDMSFSSDYSNDSWSVA-TSVT 321
Query: 449 SSPEPLPKKIRTQDQ 405
SSPEPL KKIR ++
Sbjct: 322 SSPEPLSKKIRESNE 336
>emb|CAB61223.1| cyclin D3b [Antirrhinum majus]
Length = 361
Score = 75.1 bits (183), Expect = 7e-13
Identities = 39/70 (55%), Positives = 52/70 (73%)
Frame = -1
Query: 626 DECCKVMLEMWSGIDDQGNKLCMKRKFGLIPGSPNGVVDVCFSCDSSNDSWAVAVNSVTS 447
D+C ++ E+ I++Q LC KRK+G IP SPNGV+D FS D SNDSW+ AV+SV+S
Sbjct: 287 DDCHMLITEV---INNQSYILCHKRKYGSIPSSPNGVIDAYFSSDGSNDSWS-AVSSVSS 342
Query: 446 SPEPLPKKIR 417
SPEP+ K+IR
Sbjct: 343 SPEPVFKRIR 352
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 539,160,633
Number of Sequences: 1393205
Number of extensions: 11542156
Number of successful extensions: 32223
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 31392
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32198
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25870486187
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)