Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011764A_C01 KMC011764A_c01
(718 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
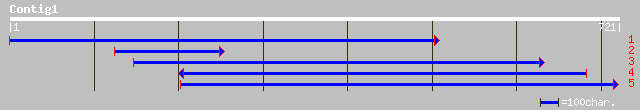
gb|AAC32170.1| GASA5-like protein [Picea mariana] gi|2996160|gb|... 128 7e-29
ref|NP_177605.1| GAST1-like protein; protein id: At1g74670.1 [Ar... 128 7e-29
pir||T51963 GASA5-like protein [imported] - Picea mariana gi|298... 127 2e-28
sp|P27057|GST1_LYCES GAST1 protein precursor gi|100217|pir||S221... 125 5e-28
emb|CAD10104.1| gibberellin induced protein 3 [Petunia x hybrida] 123 3e-27
>gb|AAC32170.1| GASA5-like protein [Picea mariana] gi|2996160|gb|AAC32171.1|
GASA5-like protein [Picea mariana]
Length = 62
Score = 128 bits (322), Expect = 7e-29
Identities = 49/61 (80%), Positives = 55/61 (89%)
Frame = -3
Query: 482 AECGPRCANRCSNTQYKKPCLFFCNKCCAKCLCVPPGLYGNKQVCPCYNNWKTKRGGPKC 303
+ECG RC+ RCS T +KKPC+FFC KCCAKCLCVPPG YGNKQVCPCYNNWKT++GGPKC
Sbjct: 2 SECGQRCSYRCSATSHKKPCMFFCQKCCAKCLCVPPGTYGNKQVCPCYNNWKTQQGGPKC 61
Query: 302 P 300
P
Sbjct: 62 P 62
>ref|NP_177605.1| GAST1-like protein; protein id: At1g74670.1 [Arabidopsis thaliana]
gi|25406361|pir||H96775 GAST1-like protein,
109761-110213 [imported] - Arabidopsis thaliana
gi|12324817|gb|AAG52379.1|AC011765_31 GAST1-like
protein; 109761-110213 [Arabidopsis thaliana]
Length = 80
Score = 128 bits (322), Expect = 7e-29
Identities = 48/60 (80%), Positives = 54/60 (90%)
Frame = -3
Query: 479 ECGPRCANRCSNTQYKKPCLFFCNKCCAKCLCVPPGLYGNKQVCPCYNNWKTKRGGPKCP 300
+CG +C RCSNT+Y KPC+FFC KCCAKCLCVPPG YGNKQVCPCYNNWKT++GGPKCP
Sbjct: 21 QCGGQCTRRCSNTKYHKPCMFFCQKCCAKCLCVPPGTYGNKQVCPCYNNWKTQQGGPKCP 80
>pir||T51963 GASA5-like protein [imported] - Picea mariana
gi|2982285|gb|AAC32128.1| GASA5-like protein [Picea
mariana]
Length = 110
Score = 127 bits (318), Expect = 2e-28
Identities = 48/61 (78%), Positives = 55/61 (89%)
Frame = -3
Query: 482 AECGPRCANRCSNTQYKKPCLFFCNKCCAKCLCVPPGLYGNKQVCPCYNNWKTKRGGPKC 303
+ECG RC+ RCS T +KKPC+FFC KCCAKCLCVPPG +GNKQVCPCYNNWKT++GGPKC
Sbjct: 50 SECGQRCSYRCSATSHKKPCMFFCQKCCAKCLCVPPGTFGNKQVCPCYNNWKTQQGGPKC 109
Query: 302 P 300
P
Sbjct: 110 P 110
>sp|P27057|GST1_LYCES GAST1 protein precursor gi|100217|pir||S22151 gibberellin-regulated
protein GAST1 - tomato gi|19247|emb|CAA44807.1| gast1
[Lycopersicon esculentum]
Length = 112
Score = 125 bits (315), Expect = 5e-28
Identities = 48/60 (80%), Positives = 51/60 (85%)
Frame = -3
Query: 479 ECGPRCANRCSNTQYKKPCLFFCNKCCAKCLCVPPGLYGNKQVCPCYNNWKTKRGGPKCP 300
+C P+C RCS T YKKPC+FFC KCCAKCLCVP G YGNKQ CPCYNNWKTKRGGPKCP
Sbjct: 53 DCQPKCTYRCSKTSYKKPCMFFCQKCCAKCLCVPAGTYGNKQSCPCYNNWKTKRGGPKCP 112
>emb|CAD10104.1| gibberellin induced protein 3 [Petunia x hybrida]
Length = 112
Score = 123 bits (308), Expect = 3e-27
Identities = 46/60 (76%), Positives = 50/60 (82%)
Frame = -3
Query: 479 ECGPRCANRCSNTQYKKPCLFFCNKCCAKCLCVPPGLYGNKQVCPCYNNWKTKRGGPKCP 300
+C P+C RCS T +KKPC+FFC KCCAKCLCVP G YGNKQ CPCYNNWKTK GGPKCP
Sbjct: 53 DCQPKCTYRCSKTSFKKPCMFFCQKCCAKCLCVPAGTYGNKQTCPCYNNWKTKEGGPKCP 112
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 635,876,769
Number of Sequences: 1393205
Number of extensions: 14177440
Number of successful extensions: 35713
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 33702
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35543
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33217548346
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)