Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011723A_C01 KMC011723A_c01
(554 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
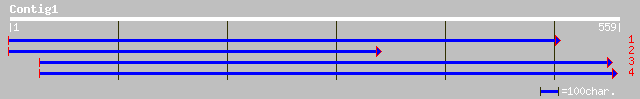
pir||T16995 probable cinnamyl-alcohol dehydrogenase (EC 1.1.1.19... 187 7e-47
pir||T11610 probable cinnamyl-alcohol dehydrogenase (EC 1.1.1.19... 184 5e-46
ref|NP_197445.1| cinnamyl-alcohol dehydrogenase - like protein; ... 173 1e-42
ref|NP_175552.1| cinnamyl alcohol dehydrogenase, putative; prote... 172 2e-42
gb|AAM65984.1| cinnamyl-alcohol dehydrogenase-like protein [Arab... 172 3e-42
>pir||T16995 probable cinnamyl-alcohol dehydrogenase (EC 1.1.1.195) - apple tree
gi|2981475|gb|AAC06319.1| putative cinnamyl alcohol
dehydrogenase [Malus x domestica]
Length = 325
Score = 187 bits (475), Expect = 7e-47
Identities = 88/139 (63%), Positives = 112/139 (80%), Gaps = 1/139 (0%)
Frame = -1
Query: 554 VVILPSMVVGPLLRPEVNFSVEPVLNIINGV-PFPNLSIGWVDVKDVAKAHIQAYEITSA 378
V I P+MV+GPLL+P +N S VLN+I G FPN S GW++VKDVA AHIQA+E +A
Sbjct: 187 VTINPAMVIGPLLQPTLNTSAAAVLNVIKGARTFPNASFGWINVKDVANAHIQAFERPTA 246
Query: 377 SGRYCLVERVAHFSEVARILHDLYPTLQISNKCVDDEPYEPTFQFSQERAKSLGIEFTPL 198
SGRYCLVERVAHFSEV RIL +LYPTLQ+ KC DD+P+ PT+Q S+E+AKSLG+EF PL
Sbjct: 247 SGRYCLVERVAHFSEVVRILRELYPTLQLPEKCADDKPFVPTYQVSKEKAKSLGVEFIPL 306
Query: 197 EISIQETVKSFREKKIINF 141
++S++ETV+S +EK +NF
Sbjct: 307 DVSLKETVESLKEKGFVNF 325
>pir||T11610 probable cinnamyl-alcohol dehydrogenase (EC 1.1.1.195) CPRD14 -
cowpea gi|1854445|dbj|BAA12161.1| CPRD14 protein [Vigna
unguiculata]
Length = 325
Score = 184 bits (468), Expect = 5e-46
Identities = 88/135 (65%), Positives = 111/135 (82%), Gaps = 1/135 (0%)
Frame = -1
Query: 542 PSMVVGPLLRPEVNFSVEPVLNIINGVP-FPNLSIGWVDVKDVAKAHIQAYEITSASGRY 366
P++VVGPLL+P +N S VLN+ING P F N+++GWVDV+DVA AH+ AYE SA+GRY
Sbjct: 191 PALVVGPLLQPVLNTSSAAVLNLINGSPTFKNVTLGWVDVRDVAIAHVLAYENASANGRY 250
Query: 365 CLVERVAHFSEVARILHDLYPTLQISNKCVDDEPYEPTFQFSQERAKSLGIEFTPLEISI 186
LVERVAHF +V +ILHDLYPTLQ+ KCVDD PY+P FQ S+E+AKSLG+EFTPLE+SI
Sbjct: 251 LLVERVAHFGDVVKILHDLYPTLQLPQKCVDDRPYDPIFQVSKEKAKSLGLEFTPLEVSI 310
Query: 185 QETVKSFREKKIINF 141
++TV+S +EK I F
Sbjct: 311 KDTVESLKEKGFIKF 325
>ref|NP_197445.1| cinnamyl-alcohol dehydrogenase - like protein; protein id:
At5g19440.1, supported by cDNA: 6748. [Arabidopsis
thaliana]
Length = 326
Score = 173 bits (439), Expect = 1e-42
Identities = 80/139 (57%), Positives = 107/139 (76%), Gaps = 1/139 (0%)
Frame = -1
Query: 554 VVILPSMVVGPLLRPEVNFSVEPVLNIINGVP-FPNLSIGWVDVKDVAKAHIQAYEITSA 378
V I P+MV+GPLL+P +N S +LN+ING FPNLS GWV+VKDVA AHIQA+E+ SA
Sbjct: 188 VTINPAMVIGPLLQPTLNTSAAAILNLINGAKTFPNLSFGWVNVKDVANAHIQAFEVPSA 247
Query: 377 SGRYCLVERVAHFSEVARILHDLYPTLQISNKCVDDEPYEPTFQFSQERAKSLGIEFTPL 198
+GRYCLVERV H SE+ IL +LYP L + +CVD+ PY PT+Q S+++ +SLGI++ PL
Sbjct: 248 NGRYCLVERVVHHSEIVNILRELYPNLPLPERCVDENPYVPTYQVSKDKTRSLGIDYIPL 307
Query: 197 EISIQETVKSFREKKIINF 141
++SI+ETV+S +EK F
Sbjct: 308 KVSIKETVESLKEKGFAQF 326
>ref|NP_175552.1| cinnamyl alcohol dehydrogenase, putative; protein id: At1g51410.1
[Arabidopsis thaliana] gi|25405464|pir||C96552
hypothetical protein F5D21.12 [imported] - Arabidopsis
thaliana gi|12325359|gb|AAG52618.1|AC024261_5 cinnamyl
alcohol dehydrogenase, putative; 82967-79323 [Arabidopsis
thaliana]
Length = 809
Score = 172 bits (436), Expect = 2e-42
Identities = 83/139 (59%), Positives = 106/139 (75%), Gaps = 1/139 (0%)
Frame = -1
Query: 554 VVILPSMVVGPLLRPEVNFSVEPVLNIINGVP-FPNLSIGWVDVKDVAKAHIQAYEITSA 378
V I P+MV+GPLL+P +N S VL++I G FPN + GWV+VKDVA AHIQA+E A
Sbjct: 671 VSINPAMVIGPLLQPTLNTSAAAVLSLIKGAQTFPNATFGWVNVKDVANAHIQAFENPDA 730
Query: 377 SGRYCLVERVAHFSEVARILHDLYPTLQISNKCVDDEPYEPTFQFSQERAKSLGIEFTPL 198
GRYCLVERVAH+SEV ILHDLYP Q+ KC D++ Y PT++ S+E+A+SLG+EF PL
Sbjct: 731 DGRYCLVERVAHYSEVVNILHDLYPDFQLPEKCADEKIYIPTYKVSKEKAESLGVEFVPL 790
Query: 197 EISIQETVKSFREKKIINF 141
E+SI+ETV+S R+K I F
Sbjct: 791 EVSIKETVESLRDKGFIRF 809
>gb|AAM65984.1| cinnamyl-alcohol dehydrogenase-like protein [Arabidopsis thaliana]
Length = 326
Score = 172 bits (435), Expect = 3e-42
Identities = 79/139 (56%), Positives = 106/139 (75%), Gaps = 1/139 (0%)
Frame = -1
Query: 554 VVILPSMVVGPLLRPEVNFSVEPVLNIINGVP-FPNLSIGWVDVKDVAKAHIQAYEITSA 378
V I P+MV+GPLL+P +N S +LN+ING FPNLS GWV+VKDVA AHIQ +E+ SA
Sbjct: 188 VTINPAMVIGPLLQPTLNTSAAAILNLINGAKTFPNLSFGWVNVKDVANAHIQTFEVPSA 247
Query: 377 SGRYCLVERVAHFSEVARILHDLYPTLQISNKCVDDEPYEPTFQFSQERAKSLGIEFTPL 198
+GRYCLVERV H SE+ IL +LYP L + +CVD+ PY PT+Q S+++ +SLGI++ PL
Sbjct: 248 NGRYCLVERVVHHSEIVNILRELYPNLPLPERCVDENPYVPTYQVSKDKTRSLGIDYIPL 307
Query: 197 EISIQETVKSFREKKIINF 141
++SI+ETV+S +EK F
Sbjct: 308 KVSIKETVESLKEKGFAQF 326
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 454,646,510
Number of Sequences: 1393205
Number of extensions: 9041733
Number of successful extensions: 21995
Number of sequences better than 10.0: 192
Number of HSP's better than 10.0 without gapping: 21338
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21839
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19521267756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)