Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011588A_C01 KMC011588A_c01
(688 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
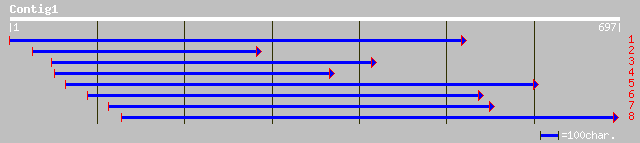
ref|NP_564010.1| expressed protein; protein id: At1g16880.1, sup... 239 3e-62
gb|AAM64912.1| unknown [Arabidopsis thaliana] 191 6e-48
ref|NP_196094.1| putative protein; protein id: At5g04740.1, supp... 191 6e-48
ref|NP_201390.1| uridylyl transferases-like; protein id: At5g658... 40 0.030
gb|AAM20640.1| translation factor EF-1 alpha-like protein [Arabi... 40 0.039
>ref|NP_564010.1| expressed protein; protein id: At1g16880.1, supported by cDNA:
gi_14423501 [Arabidopsis thaliana]
gi|25344189|pir||B86304 hypothetical protein F6I1.12
[imported] - Arabidopsis thaliana
gi|9802776|gb|AAF99845.1|AC051629_12 Unknown protein
[Arabidopsis thaliana]
gi|14423502|gb|AAK62433.1|AF386988_1 Unknown protein
[Arabidopsis thaliana]
Length = 290
Score = 239 bits (610), Expect = 3e-62
Identities = 110/144 (76%), Positives = 139/144 (96%)
Frame = -3
Query: 686 TGRKVDDPELLEAIRMTILNNMIQYHPESSAQLALGAAFGLVPPKEQVDVDIATHLTISD 507
+GRKV+DPELLEAIR+T++NN++++HPESS+QLA+GAAFG++PP E +DVDIATH+TI D
Sbjct: 147 SGRKVEDPELLEAIRLTVINNLLEFHPESSSQLAMGAAFGVLPPTEPIDVDIATHITIED 206
Query: 506 DGPDRSLLYVETADRPGLLLDLVQIITDINIAVESGEFDTEGLLAKAKFHVSYKGKAIIK 327
DGPDRSLL++E+ADRPGLL++LV+II+DI++AVESGEFDTEGLLAK KFHVSY+ KA+IK
Sbjct: 207 DGPDRSLLFIESADRPGLLVELVKIISDISVAVESGEFDTEGLLAKVKFHVSYRNKALIK 266
Query: 326 PLQEVLANSLRYYLRRPSTEESSF 255
PLQ+VLANSLRY+LRRPST+ESSF
Sbjct: 267 PLQQVLANSLRYFLRRPSTDESSF 290
>gb|AAM64912.1| unknown [Arabidopsis thaliana]
Length = 301
Score = 191 bits (486), Expect = 6e-48
Identities = 85/144 (59%), Positives = 120/144 (83%)
Frame = -3
Query: 686 TGRKVDDPELLEAIRMTILNNMIQYHPESSAQLALGAAFGLVPPKEQVDVDIATHLTISD 507
TGRKV+DP+LLE IR+TI+NN+++YHPE S QLA+G FG+ P++++DVDIATH+ + +
Sbjct: 158 TGRKVEDPDLLEQIRLTIINNLLKYHPECSEQLAMGETFGIKAPEKKIDVDIATHIHVKE 217
Query: 506 DGPDRSLLYVETADRPGLLLDLVQIITDINIAVESGEFDTEGLLAKAKFHVSYKGKAIIK 327
DGP RSLL +ETADRPGL++++++++ D+NI VES E DTEGL+AK KFHVSY+G+A+ +
Sbjct: 218 DGPKRSLLVIETADRPGLVVEMIKVMADVNIDVESAEIDTEGLVAKDKFHVSYQGQALNR 277
Query: 326 PLQEVLANSLRYYLRRPSTEESSF 255
L +VL N LRY+LRRP T+ S+
Sbjct: 278 SLSQVLVNCLRYFLRRPETDIDSY 301
Score = 37.4 bits (85), Expect = 0.20
Identities = 30/128 (23%), Positives = 61/128 (47%), Gaps = 10/128 (7%)
Frame = -3
Query: 641 MTILNNMIQYHPESSAQLALGAAFGLVPPKEQVD--VDIATHLTISDDGPDRSLLYVETA 468
M++L I+ +S AA P E D V + + D P+ +++ +
Sbjct: 56 MSLLTKSIKNRVYASINSIDSAATPSYPKSEDDDDVVPMPMVMIDQDADPEATIVQLSFG 115
Query: 467 DRPGLLLDLVQIITDINIAVESGEFDTEGLLAKAKFHVSYK--GKAIIKP--LQE----V 312
+R G L+D ++ + D+ + V G TEG + + KF ++ + G+ + P L++ +
Sbjct: 116 NRLGALIDTMRALKDLGLDVIKGTVSTEGSIKQTKFSITKRDTGRKVEDPDLLEQIRLTI 175
Query: 311 LANSLRYY 288
+ N L+Y+
Sbjct: 176 INNLLKYH 183
>ref|NP_196094.1| putative protein; protein id: At5g04740.1, supported by cDNA:
34592., supported by cDNA: gi_18252932 [Arabidopsis
thaliana] gi|11281139|pir||T48470 hypothetical protein
T1E3.100 - Arabidopsis thaliana
gi|7413536|emb|CAB86016.1| putative protein [Arabidopsis
thaliana] gi|9758449|dbj|BAB08978.1|
gene_id:MUK11.6~unknown protein [Arabidopsis thaliana]
gi|18252933|gb|AAL62393.1| putative protein [Arabidopsis
thaliana] gi|21389645|gb|AAM48021.1| putative protein
[Arabidopsis thaliana]
Length = 301
Score = 191 bits (486), Expect = 6e-48
Identities = 85/144 (59%), Positives = 120/144 (83%)
Frame = -3
Query: 686 TGRKVDDPELLEAIRMTILNNMIQYHPESSAQLALGAAFGLVPPKEQVDVDIATHLTISD 507
TGRKV+DP+LLE IR+TI+NN+++YHPE S QLA+G FG+ P++++DVDIATH+ + +
Sbjct: 158 TGRKVEDPDLLEQIRLTIINNLLKYHPECSEQLAMGETFGIKAPEKKIDVDIATHIHVKE 217
Query: 506 DGPDRSLLYVETADRPGLLLDLVQIITDINIAVESGEFDTEGLLAKAKFHVSYKGKAIIK 327
DGP RSLL +ETADRPGL++++++++ D+NI VES E DTEGL+AK KFHVSY+G+A+ +
Sbjct: 218 DGPKRSLLVIETADRPGLVVEMIKVMADVNIDVESAEIDTEGLVAKDKFHVSYQGQALNR 277
Query: 326 PLQEVLANSLRYYLRRPSTEESSF 255
L +VL N LRY+LRRP T+ S+
Sbjct: 278 SLSQVLVNCLRYFLRRPETDIDSY 301
Score = 37.4 bits (85), Expect = 0.20
Identities = 30/128 (23%), Positives = 61/128 (47%), Gaps = 10/128 (7%)
Frame = -3
Query: 641 MTILNNMIQYHPESSAQLALGAAFGLVPPKEQVD--VDIATHLTISDDGPDRSLLYVETA 468
M++L I+ +S AA P E D V + + D P+ +++ +
Sbjct: 56 MSLLTKSIKNRVYASINSIDSAATPSYPKSEDDDDVVPMPMVMIDQDADPEATIVQLSFG 115
Query: 467 DRPGLLLDLVQIITDINIAVESGEFDTEGLLAKAKFHVSYK--GKAIIKP--LQE----V 312
+R G L+D ++ + D+ + V G TEG + + KF ++ + G+ + P L++ +
Sbjct: 116 NRLGALIDTMRALKDLGLDVIKGTVSTEGSIKQTKFSITKRDTGRKVEDPDLLEQIRLTI 175
Query: 311 LANSLRYY 288
+ N L+Y+
Sbjct: 176 INNLLKYH 183
>ref|NP_201390.1| uridylyl transferases-like; protein id: At5g65890.1, supported by
cDNA: gi_13430687 [Arabidopsis thaliana]
gi|9759573|dbj|BAB11136.1| uridylyl transferases-like
[Arabidopsis thaliana]
gi|13430688|gb|AAK25966.1|AF360256_1 putative uridylyl
transferases [Arabidopsis thaliana]
gi|14532892|gb|AAK64128.1| putative uridylyl
transferases [Arabidopsis thaliana]
gi|22138094|gb|AAM93427.1| ACR1 [Arabidopsis thaliana]
Length = 477
Score = 40.0 bits (92), Expect = 0.030
Identities = 20/70 (28%), Positives = 40/70 (56%)
Frame = -3
Query: 560 PPKEQVDVDIATHLTISDDGPDRSLLYVETADRPGLLLDLVQIITDINIAVESGEFDTEG 381
PP+ VD +D P+ +L+ V++A++ G+LLD+VQ++ D+++ + ++G
Sbjct: 23 PPRVCVD---------NDSDPECTLIKVDSANKYGILLDMVQVLADLDLVISKCYISSDG 73
Query: 380 LLAKAKFHVS 351
FHV+
Sbjct: 74 EWFMDVFHVT 83
>gb|AAM20640.1| translation factor EF-1 alpha-like protein [Arabidopsis thaliana]
Length = 449
Score = 39.7 bits (91), Expect = 0.039
Identities = 18/57 (31%), Positives = 37/57 (64%), Gaps = 2/57 (3%)
Frame = -3
Query: 515 ISDDG--PDRSLLYVETADRPGLLLDLVQIITDINIAVESGEFDTEGLLAKAKFHVS 351
+ D+G P+ +++ +++A PG+LL+ VQ++TD+N+ ++ ++G FHVS
Sbjct: 23 VIDNGVCPNSTVVKIDSARSPGILLESVQLLTDMNLWIKKAYISSDGKWNMDVFHVS 79
Score = 34.7 bits (78), Expect = 1.3
Identities = 16/50 (32%), Positives = 29/50 (58%)
Frame = -3
Query: 473 TADRPGLLLDLVQIITDINIAVESGEFDTEGLLAKAKFHVSYKGKAIIKP 324
TADRPGLL ++ +I+ + + + E T+ +A+ F+V+ +I P
Sbjct: 329 TADRPGLLAEVTRILRENGLNIARAEISTKDSIARNVFYVTDANGNLIDP 378
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 581,380,089
Number of Sequences: 1393205
Number of extensions: 12177740
Number of successful extensions: 30981
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 29947
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30937
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30835865868
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)