Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011562A_C01 KMC011562A_c01
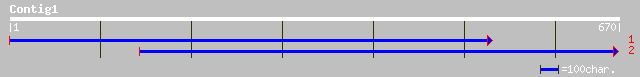
(670 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_567387.1| putative protein; protein id: At4g12070.1, supp... 224 1e-57
gb|AAM64931.1| unknown [Arabidopsis thaliana] 223 2e-57
pir||T06614 hypothetical protein F16J13.140 - Arabidopsis thalia... 211 7e-54
emb|CAC28699.2| hypothetical protein [Neurospora crassa] 36 0.41
dbj|BAC65530.1| mKIAA0324 protein [Mus musculus] 35 0.92
>ref|NP_567387.1| putative protein; protein id: At4g12070.1, supported by cDNA:
34781., supported by cDNA: gi_14994258 [Arabidopsis
thaliana] gi|14994259|gb|AAK73264.1| putative protein
[Arabidopsis thaliana] gi|26451708|dbj|BAC42949.1|
unknown protein [Arabidopsis thaliana]
Length = 483
Score = 224 bits (570), Expect = 1e-57
Identities = 110/182 (60%), Positives = 139/182 (75%), Gaps = 7/182 (3%)
Frame = +1
Query: 145 QTHRHETPKSYPS-------ECLRGSSKADEWTGDMLQTGDIVEELRIGGSVNSLIRFKS 303
+TH+ E+P + P +CL+GSSK++EW+GDMLQTGDIVEE+RIG S I FK+
Sbjct: 3 RTHQPESPTARPGNTVVLSIDCLKGSSKSEEWSGDMLQTGDIVEEIRIGSGPGSAI-FKA 61
Query: 304 PFKSGKSGVNKILQDAYKKKETSICVRVRRGPDLFAEWQACIVPNESSGKKNYVLRSISD 483
PFK GK+ + K+L ++++ KETSI VRVRRG D F++ ACIVPNES+GK+ Y+LRSI D
Sbjct: 62 PFKGGKAWLQKVLHNSFRNKETSIVVRVRRGSDDFSDLSACIVPNESAGKRQYMLRSIDD 121
Query: 484 PNYVVGFLDRTEAECFQLQASRSSRMVHALTKTRLQDGFVSYPWERRMHE*LSVPNTSNF 663
PNY VGF DRTE++C LQ SR SRMV AL K +LQDG+VSYPWERRM E L + +SNF
Sbjct: 122 PNYTVGFSDRTESDCLGLQESRGSRMVEALVKAKLQDGYVSYPWERRMQEALPISGSSNF 181
Query: 664 LS 669
LS
Sbjct: 182 LS 183
>gb|AAM64931.1| unknown [Arabidopsis thaliana]
Length = 483
Score = 223 bits (567), Expect = 2e-57
Identities = 109/182 (59%), Positives = 139/182 (75%), Gaps = 7/182 (3%)
Frame = +1
Query: 145 QTHRHETPKSYPS-------ECLRGSSKADEWTGDMLQTGDIVEELRIGGSVNSLIRFKS 303
+TH+ E+P + P +CL+GSSK++EW+GDMLQTGDIVEE+RIG S I FK+
Sbjct: 3 RTHQPESPTARPGNTVVLSIDCLKGSSKSEEWSGDMLQTGDIVEEIRIGSGPGSAI-FKA 61
Query: 304 PFKSGKSGVNKILQDAYKKKETSICVRVRRGPDLFAEWQACIVPNESSGKKNYVLRSISD 483
PFK GK+ + K+L ++++ KETSI VRVRRG D F++ ACIVPNES+GK+ Y+LRSI D
Sbjct: 62 PFKGGKAWLQKVLHNSFRNKETSIVVRVRRGSDDFSDLSACIVPNESAGKRQYMLRSIDD 121
Query: 484 PNYVVGFLDRTEAECFQLQASRSSRMVHALTKTRLQDGFVSYPWERRMHE*LSVPNTSNF 663
PNY VGF DRTE++C LQ SR SRMV AL K +LQDG+VSYPWE+RM E L + +SNF
Sbjct: 122 PNYTVGFSDRTESDCLGLQESRGSRMVEALVKAKLQDGYVSYPWEKRMQEALPISGSSNF 181
Query: 664 LS 669
LS
Sbjct: 182 LS 183
>pir||T06614 hypothetical protein F16J13.140 - Arabidopsis thaliana
gi|4586112|emb|CAB40948.1| putative protein [Arabidopsis
thaliana] gi|7267908|emb|CAB78250.1| putative protein
[Arabidopsis thaliana]
Length = 501
Score = 211 bits (537), Expect = 7e-54
Identities = 110/205 (53%), Positives = 139/205 (67%), Gaps = 30/205 (14%)
Frame = +1
Query: 145 QTHRHETPKSYPS-------ECLRGSSKADEWTGDMLQTGDIVEELRIGGSVNSLIRFKS 303
+TH+ E+P + P +CL+GSSK++EW+GDMLQTGDIVEE+RIG S I FK+
Sbjct: 3 RTHQPESPTARPGNTVVLSIDCLKGSSKSEEWSGDMLQTGDIVEEIRIGSGPGSAI-FKA 61
Query: 304 PFKSGKSGVNKILQDAYKKKETSICVRVRRGPDLFAEWQACIVPNESSGKKNYVLRSISD 483
PFK GK+ + K+L ++++ KETSI VRVRRG D F++ ACIVPNES+GK+ Y+LRSI D
Sbjct: 62 PFKGGKAWLQKVLHNSFRNKETSIVVRVRRGSDDFSDLSACIVPNESAGKRQYMLRSIDD 121
Query: 484 PNYVVGFLDRTEAECFQLQA-----------------------SRSSRMVHALTKTRLQD 594
PNY VGF DRTE++C LQ SR SRMV AL K +LQD
Sbjct: 122 PNYTVGFSDRTESDCLGLQVGEKSINVVFLNKCDFIVDQKNIESRGSRMVEALVKAKLQD 181
Query: 595 GFVSYPWERRMHE*LSVPNTSNFLS 669
G+VSYPWERRM E L + +SNFLS
Sbjct: 182 GYVSYPWERRMQEALPISGSSNFLS 206
>emb|CAC28699.2| hypothetical protein [Neurospora crassa]
Length = 435
Score = 36.2 bits (82), Expect = 0.41
Identities = 33/124 (26%), Positives = 51/124 (40%), Gaps = 6/124 (4%)
Frame = +2
Query: 101 HHQEHDHGQSLTKQFRLTDTRHQSPIHLSVFEEAPKPTNGPATCSKP-AT*SKSFESAVR 277
H Q+H H K+ + D+ H+ P+ S+F K GP P A+ +S
Sbjct: 258 HSQDHHHH----KRTKSHDSNHKQPLLFSIFYAVTKTHAGPQIRRMPSASPHRSSTRGKS 313
Query: 278 LTR-----SSGSSRRSRAARAASTRSFRMRIRRRKPPSVSVSAVDLTCSPNGRLVSFLMN 442
TR S+ +S S A +AS R + K P V ++ P V ++
Sbjct: 314 TTRTSTRPSTATSSSSSRAPSASKPPVTARPTKPKKPPVPTASETTVVVPLDTGVPSVVA 373
Query: 443 PPGR 454
PPG+
Sbjct: 374 PPGK 377
>dbj|BAC65530.1| mKIAA0324 protein [Mus musculus]
Length = 1754
Score = 35.0 bits (79), Expect = 0.92
Identities = 22/54 (40%), Positives = 30/54 (54%)
Frame = +2
Query: 221 PATCSKPAT*SKSFESAVRLTRSSGSSRRSRAARAASTRSFRMRIRRRKPPSVS 382
P T S+ +S S+ LTR + SRRSR+A ++ R RRR+ PSVS
Sbjct: 685 PKTKSRTPPRRRSSRSSPELTRKARVSRRSRSASSSPEIRSRTPPRRRRSPSVS 738
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 588,772,618
Number of Sequences: 1393205
Number of extensions: 13072208
Number of successful extensions: 42440
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 40211
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 42338
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29138478756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)