Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011159A_C01 KMC011159A_c01
(716 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
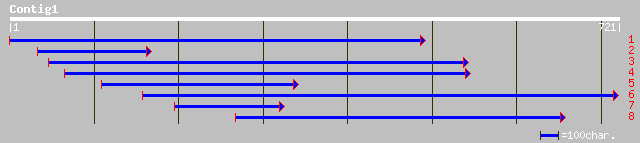
gb|AAM18095.1|AF498303_1 putative sodium-dependent bile acid sym... 197 1e-49
ref|NP_180258.2| putative Na+ dependent ileal bile acid transpor... 197 1e-49
gb|AAO41972.1| putative Na+ dependent ileal bile acid transporte... 197 1e-49
dbj|BAB40160.1| P0707D10.18 [Oryza sativa (japonica cultivar-gro... 184 1e-45
pir||T02645 hypothetical protein At2g26900 [imported] - Arabidop... 151 8e-36
>gb|AAM18095.1|AF498303_1 putative sodium-dependent bile acid symporter [Arabidopsis
thaliana]
Length = 409
Score = 197 bits (501), Expect = 1e-49
Identities = 98/109 (89%), Positives = 101/109 (91%)
Frame = -2
Query: 715 PIGQVAEVLKTQGAQLILPVMALHAAAFALGYWMSKLSFGESTSRPKSIECGMQSSALGF 536
PIGQVA+VLKTQGAQLILPV LHAAAFA+GYW+SK SFGESTSR SIECGMQSSALGF
Sbjct: 301 PIGQVADVLKTQGAQLILPVALLHAAAFAIGYWISKFSFGESTSRTISIECGMQSSALGF 360
Query: 535 LLAQKHFTNPLVAVPSAVSVVCMALGGSALAVFWRNSPIPIDDKDDFKE 389
LLAQKHFTNPLVAVPSAVSVVCMALGGS LAVFWRN PIP DDKDDFKE
Sbjct: 361 LLAQKHFTNPLVAVPSAVSVVCMALGGSGLAVFWRNLPIPADDKDDFKE 409
>ref|NP_180258.2| putative Na+ dependent ileal bile acid transporter; protein id:
At2g26900.1, supported by cDNA: gi_20269900 [Arabidopsis
thaliana]
Length = 409
Score = 197 bits (501), Expect = 1e-49
Identities = 98/109 (89%), Positives = 101/109 (91%)
Frame = -2
Query: 715 PIGQVAEVLKTQGAQLILPVMALHAAAFALGYWMSKLSFGESTSRPKSIECGMQSSALGF 536
PIGQVA+VLKTQGAQLILPV LHAAAFA+GYW+SK SFGESTSR SIECGMQSSALGF
Sbjct: 301 PIGQVADVLKTQGAQLILPVALLHAAAFAIGYWISKFSFGESTSRTISIECGMQSSALGF 360
Query: 535 LLAQKHFTNPLVAVPSAVSVVCMALGGSALAVFWRNSPIPIDDKDDFKE 389
LLAQKHFTNPLVAVPSAVSVVCMALGGS LAVFWRN PIP DDKDDFKE
Sbjct: 361 LLAQKHFTNPLVAVPSAVSVVCMALGGSGLAVFWRNLPIPADDKDDFKE 409
>gb|AAO41972.1| putative Na+ dependent ileal bile acid transporter [Arabidopsis
thaliana]
Length = 271
Score = 197 bits (501), Expect = 1e-49
Identities = 98/109 (89%), Positives = 101/109 (91%)
Frame = -2
Query: 715 PIGQVAEVLKTQGAQLILPVMALHAAAFALGYWMSKLSFGESTSRPKSIECGMQSSALGF 536
PIGQVA+VLKTQGAQLILPV LHAAAFA+GYW+SK SFGESTSR SIECGMQSSALGF
Sbjct: 163 PIGQVADVLKTQGAQLILPVALLHAAAFAIGYWISKFSFGESTSRTISIECGMQSSALGF 222
Query: 535 LLAQKHFTNPLVAVPSAVSVVCMALGGSALAVFWRNSPIPIDDKDDFKE 389
LLAQKHFTNPLVAVPSAVSVVCMALGGS LAVFWRN PIP DDKDDFKE
Sbjct: 223 LLAQKHFTNPLVAVPSAVSVVCMALGGSGLAVFWRNLPIPADDKDDFKE 271
>dbj|BAB40160.1| P0707D10.18 [Oryza sativa (japonica cultivar-group)]
Length = 408
Score = 184 bits (467), Expect = 1e-45
Identities = 93/110 (84%), Positives = 99/110 (89%), Gaps = 1/110 (0%)
Frame = -2
Query: 715 PIGQVAEVLKTQGAQLILPVMALHAAAFALGYWMSKLS-FGESTSRPKSIECGMQSSALG 539
PIGQV+EVLK QG QLI+PV LH AAFALGYW+SK+S FGESTSR SIECGMQSSALG
Sbjct: 299 PIGQVSEVLKAQGGQLIIPVALLHVAAFALGYWLSKVSSFGESTSRTISIECGMQSSALG 358
Query: 538 FLLAQKHFTNPLVAVPSAVSVVCMALGGSALAVFWRNSPIPIDDKDDFKE 389
FLLAQKHFTNPLVAVPSAVSVVCMALGGSALAVFWRN +P +DKDDFKE
Sbjct: 359 FLLAQKHFTNPLVAVPSAVSVVCMALGGSALAVFWRNRGLPANDKDDFKE 408
>pir||T02645 hypothetical protein At2g26900 [imported] - Arabidopsis thaliana
gi|3426051|gb|AAC32250.1| putative Na+ dependent ileal
bile acid transporter [Arabidopsis thaliana]
Length = 338
Score = 151 bits (382), Expect = 8e-36
Identities = 76/85 (89%), Positives = 80/85 (93%)
Frame = -2
Query: 715 PIGQVAEVLKTQGAQLILPVMALHAAAFALGYWMSKLSFGESTSRPKSIECGMQSSALGF 536
PIGQVA+VLKTQGAQLILPV LHAAAFA+GYW+SK SFGESTSR SIECGMQSSALGF
Sbjct: 240 PIGQVADVLKTQGAQLILPVALLHAAAFAIGYWISKFSFGESTSRTISIECGMQSSALGF 299
Query: 535 LLAQKHFTNPLVAVPSAVSVVCMAL 461
LLAQKHFTNPLVAVPSAVSVVCMA+
Sbjct: 300 LLAQKHFTNPLVAVPSAVSVVCMAV 324
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 624,460,391
Number of Sequences: 1393205
Number of extensions: 13820086
Number of successful extensions: 35129
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 33313
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35009
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33217548346
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)