Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011135A_C01 KMC011135A_c01
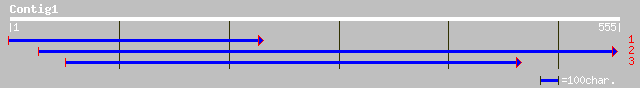
(554 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB09194.1| contains similarity to unknown protein~dbj|BAA76... 113 2e-24
ref|NP_568616.1| putative protein; protein id: At5g42920.1, supp... 113 2e-24
ref|NP_683401.1| unknown protein; protein id: At1g45250.1 [Arabi... 100 9e-21
ref|NP_035251.1| polo-like kinase homolog; polo-like kinase homo... 32 4.0
ref|NP_058796.1| polo-like kinase homolog [Rattus norvegicus] gi... 32 4.0
>dbj|BAB09194.1| contains similarity to unknown
protein~dbj|BAA76827.1~gene_id:MBD2.12 [Arabidopsis
thaliana]
Length = 824
Score = 113 bits (282), Expect = 2e-24
Identities = 56/113 (49%), Positives = 75/113 (65%), Gaps = 3/113 (2%)
Frame = -3
Query: 552 EGSDDGTDNDILCNLFPNDTGLELPHQSAQLFVGDAVTFNAQRASRPYKWAQHLAGTDFL 373
EGS DG + +I CNLFP+D GLE PHQS +L +GD TF+ R SRPYKW QHLAG D
Sbjct: 380 EGSQDGPEKNIFCNLFPDDAGLEPPHQSTKLILGDGQTFDENRTSRPYKWVQHLAGID-- 437
Query: 372 PEVSPLLLND---NSEATTTEDVISGLSLYRHTNRVQTLLHRTSLKEKSSVGL 223
+SP+LL N++ ++ + LSLYR +RVQT+L R L++K+ + L
Sbjct: 438 --ISPVLLGQEAHNTDPAKSDTFVPDLSLYRQQHRVQTVLRRIRLRKKAHLAL 488
>ref|NP_568616.1| putative protein; protein id: At5g42920.1, supported by cDNA:
gi_14532601, supported by cDNA: gi_20259138 [Arabidopsis
thaliana] gi|14532602|gb|AAK64029.1| unknown protein
[Arabidopsis thaliana] gi|20259139|gb|AAM14285.1|
unknown protein [Arabidopsis thaliana]
Length = 702
Score = 113 bits (282), Expect = 2e-24
Identities = 56/113 (49%), Positives = 75/113 (65%), Gaps = 3/113 (2%)
Frame = -3
Query: 552 EGSDDGTDNDILCNLFPNDTGLELPHQSAQLFVGDAVTFNAQRASRPYKWAQHLAGTDFL 373
EGS DG + +I CNLFP+D GLE PHQS +L +GD TF+ R SRPYKW QHLAG D
Sbjct: 263 EGSQDGPEKNIFCNLFPDDAGLEPPHQSTKLILGDGQTFDENRTSRPYKWVQHLAGID-- 320
Query: 372 PEVSPLLLND---NSEATTTEDVISGLSLYRHTNRVQTLLHRTSLKEKSSVGL 223
+SP+LL N++ ++ + LSLYR +RVQT+L R L++K+ + L
Sbjct: 321 --ISPVLLGQEAHNTDPAKSDTFVPDLSLYRQQHRVQTVLRRIRLRKKAHLAL 371
>ref|NP_683401.1| unknown protein; protein id: At1g45250.1 [Arabidopsis thaliana]
Length = 452
Score = 100 bits (250), Expect = 9e-21
Identities = 51/111 (45%), Positives = 75/111 (66%)
Frame = -3
Query: 552 EGSDDGTDNDILCNLFPNDTGLELPHQSAQLFVGDAVTFNAQRASRPYKWAQHLAGTDFL 373
E S+DG + +ILCNLFP+D+GLE PHQSA+L +G+ F+ R SRPYKWAQHLAG + L
Sbjct: 345 EESEDGLEKNILCNLFPDDSGLEPPHQSAKLILGNDHVFDKSRTSRPYKWAQHLAGIETL 404
Query: 372 PEVSPLLLNDNSEATTTEDVISGLSLYRHTNRVQTLLHRTSLKEKSSVGLL 220
PE+SP + + + + T + S +R+ VQT+L R ++K+ + L+
Sbjct: 405 PEMSPFFTDKDIQYSDTAKGYASASDHRN---VQTVLQRIRSQKKTKLTLV 452
>ref|NP_035251.1| polo-like kinase homolog; polo-like kinase homolog, (Drosophila)
[Mus musculus] gi|1709659|sp|Q07832|PLK1_MOUSE
Serine/threonine-protein kinase PLK (PLK-1)
(Serine-threonine protein kinase 13) (STPK13)
gi|2137690|pir||A54596 protein kinase - mouse
gi|393038|gb|AAA56635.1| pLK gi|403474|gb|AAA16071.1|
protein kinase gi|13905176|gb|AAH06880.1| polo-like
kinase homolog, (Drosophila) [Mus musculus]
Length = 603
Score = 32.3 bits (72), Expect = 4.0
Identities = 19/61 (31%), Positives = 35/61 (57%)
Frame = +3
Query: 135 SNYRFIVIEYKKRS*IF*NHKKARALTHTRGQLSFFP*AKFYVEVSVLGSCDDIVKDQIL 314
S++ F+V+E +R + HK+ +ALT P A++Y+ VLG C + ++Q++
Sbjct: 123 SDFVFVVLELCRRRSLLELHKRRKALTE--------PEARYYLRQIVLG-CQYLHRNQVI 173
Query: 315 H 317
H
Sbjct: 174 H 174
>ref|NP_058796.1| polo-like kinase homolog [Rattus norvegicus]
gi|12230396|sp|Q62673|PLK1_RAT Serine/threonine-protein
kinase PLK (PLK-1) gi|497994|gb|AAA18885.1| polo like
kinase
Length = 603
Score = 32.3 bits (72), Expect = 4.0
Identities = 19/61 (31%), Positives = 35/61 (57%)
Frame = +3
Query: 135 SNYRFIVIEYKKRS*IF*NHKKARALTHTRGQLSFFP*AKFYVEVSVLGSCDDIVKDQIL 314
S++ F+V+E +R + HK+ +ALT P A++Y+ VLG C + ++Q++
Sbjct: 123 SDFVFVVLELCRRRSLLELHKRRKALTE--------PEARYYLRQIVLG-CQYLHRNQVI 173
Query: 315 H 317
H
Sbjct: 174 H 174
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 424,402,506
Number of Sequences: 1393205
Number of extensions: 8224967
Number of successful extensions: 23847
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 23299
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23844
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19521267756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)