Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011123A_C01 KMC011123A_c01
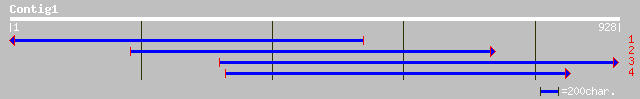
(928 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAC97389.1| 4-coumarate:CoA ligase isoenzyme 3 [Glycine max] ... 525 e-148
pir||PQ0772 4-coumarate-CoA ligase (EC 6.2.1.12) (clone GM4CL1B)... 523 e-147
sp|P31687|4CL2_SOYBN 4-coumarate--CoA ligase 2 (4CL 2) (4-coumar... 523 e-147
pir||T08075 4-coumarate-CoA ligase (EC 6.2.1.12) (clone 4CL2) [v... 476 e-133
gb|AAK58909.1| 4-coumarate:CoA ligase 4 [Populus balsamifera sub... 473 e-132
>gb|AAC97389.1| 4-coumarate:CoA ligase isoenzyme 3 [Glycine max]
gi|4038973|gb|AAC97599.1| 4-coumarate:CoA ligase
isoenzyme 3 [Glycine max]
Length = 570
Score = 525 bits (1351), Expect = e-148
Identities = 267/309 (86%), Positives = 284/309 (91%)
Frame = -2
Query: 927 KLVITQAMYVDKLRPLEDVNGNDYPKLGEDFKVVTVDEQPKDCLDFSVISEGKEDDLPEV 748
KL+ITQAMYVDKLR N +D KLGEDFKVVTVD+ P++CL FSV+SE E D PEV
Sbjct: 148 KLIITQAMYVDKLR-----NHDDGAKLGEDFKVVTVDDPPENCLHFSVLSEANESDAPEV 202
Query: 747 EINPEDAVALPFSSGTTGLPKGVVLTHKSLTTSVGQQVDGENPNLSLGTEDVLLCVLPLF 568
+I P+DAVA+PFSSGTTGLPKGVVLTHKSLTTSV QQVDGENPNL L TEDVLLCVLPLF
Sbjct: 203 DIQPDDAVAMPFSSGTTGLPKGVVLTHKSLTTSVAQQVDGENPNLYLTTEDVLLCVLPLF 262
Query: 567 HIFSLNSVLLCALRAGSGVLLMPKFEIGSLLELIQKHRVSVAMVVPPLVLALAKNPKVAE 388
HIFSLNSVLLCALRAGS VLLM KFEIG+LLELIQ+HRVSVAMVVPPLVLALAKNP VA+
Sbjct: 263 HIFSLNSVLLCALRAGSAVLLMQKFEIGTLLELIQRHRVSVAMVVPPLVLALAKNPMVAD 322
Query: 387 FDLSSIRLVLSGAAPLGKELEEALRSRVPQAVLGQGYGMTEAGPVLSMSLGFAKQPFPTK 208
FDLSSIRLVLSGAAPLGKEL EALR+RVPQAVLGQGYGMTEAGPVLSM LGFAKQPFPTK
Sbjct: 323 FDLSSIRLVLSGAAPLGKELVEALRNRVPQAVLGQGYGMTEAGPVLSMCLGFAKQPFPTK 382
Query: 207 SGSCGTVVRNAELKVLDPETGCSLGYNQPGEICIRGQQIMKGYLNDEKATATTIDAEGWL 28
SGSCGTVVRNAEL+V+DPETG SLGYNQPGEICIRGQQIMKGYLNDEKATA TID+EGWL
Sbjct: 383 SGSCGTVVRNAELRVVDPETGRSLGYNQPGEICIRGQQIMKGYLNDEKATALTIDSEGWL 442
Query: 27 HTGDVGYID 1
HTGDVGY+D
Sbjct: 443 HTGDVGYVD 451
>pir||PQ0772 4-coumarate-CoA ligase (EC 6.2.1.12) (clone GM4CL1B) - soybean
(fragment)
Length = 423
Score = 523 bits (1346), Expect = e-147
Identities = 265/309 (85%), Positives = 285/309 (91%)
Frame = -2
Query: 927 KLVITQAMYVDKLRPLEDVNGNDYPKLGEDFKVVTVDEQPKDCLDFSVISEGKEDDLPEV 748
KL+ITQAMYVDKLR +D KLGEDFKVVTVD+ P++CL FSV+SE E D+PEV
Sbjct: 2 KLIITQAMYVDKLR------NHDGAKLGEDFKVVTVDDPPENCLHFSVLSEANESDVPEV 55
Query: 747 EINPEDAVALPFSSGTTGLPKGVVLTHKSLTTSVGQQVDGENPNLSLGTEDVLLCVLPLF 568
EI+P+DAVA+PFSSGTTGLPKGV+LTHKSLTTSV QQVDGENPNL L TEDVLLCVLPLF
Sbjct: 56 EIHPDDAVAMPFSSGTTGLPKGVILTHKSLTTSVAQQVDGENPNLYLTTEDVLLCVLPLF 115
Query: 567 HIFSLNSVLLCALRAGSGVLLMPKFEIGSLLELIQKHRVSVAMVVPPLVLALAKNPKVAE 388
HIFSLNSVLLCALRAGS VLLM KFEIG+LLELIQ+HRVSVAMVVPPLVLALAKNP VA+
Sbjct: 116 HIFSLNSVLLCALRAGSAVLLMQKFEIGTLLELIQRHRVSVAMVVPPLVLALAKNPMVAD 175
Query: 387 FDLSSIRLVLSGAAPLGKELEEALRSRVPQAVLGQGYGMTEAGPVLSMSLGFAKQPFPTK 208
FDLSSIRLVLSGAAPLGKELEEALR+R+PQAVLGQGYGMTEAGPVLSM LGFAKQPF TK
Sbjct: 176 FDLSSIRLVLSGAAPLGKELEEALRNRMPQAVLGQGYGMTEAGPVLSMCLGFAKQPFQTK 235
Query: 207 SGSCGTVVRNAELKVLDPETGCSLGYNQPGEICIRGQQIMKGYLNDEKATATTIDAEGWL 28
SGSCGTVVRNAELKV+DPETG SLGYNQPGEICIRGQQIMKGYLNDE ATA+TID+EGWL
Sbjct: 236 SGSCGTVVRNAELKVVDPETGRSLGYNQPGEICIRGQQIMKGYLNDEAATASTIDSEGWL 295
Query: 27 HTGDVGYID 1
HTGDVGY+D
Sbjct: 296 HTGDVGYVD 304
>sp|P31687|4CL2_SOYBN 4-coumarate--CoA ligase 2 (4CL 2) (4-coumaroyl-CoA synthase 2) (Clone
4CL16) gi|13559169|emb|CAC36095.1| 4-coumarate:Coenzyme A
ligase isoenzyme 4 [Glycine max]
Length = 562
Score = 523 bits (1346), Expect = e-147
Identities = 265/309 (85%), Positives = 285/309 (91%)
Frame = -2
Query: 927 KLVITQAMYVDKLRPLEDVNGNDYPKLGEDFKVVTVDEQPKDCLDFSVISEGKEDDLPEV 748
KL+ITQAMYVDKLR +D KLGEDFKVVTVD+ P++CL FSV+SE E D+PEV
Sbjct: 141 KLIITQAMYVDKLR------NHDGAKLGEDFKVVTVDDPPENCLHFSVLSEANESDVPEV 194
Query: 747 EINPEDAVALPFSSGTTGLPKGVVLTHKSLTTSVGQQVDGENPNLSLGTEDVLLCVLPLF 568
EI+P+DAVA+PFSSGTTGLPKGV+LTHKSLTTSV QQVDGENPNL L TEDVLLCVLPLF
Sbjct: 195 EIHPDDAVAMPFSSGTTGLPKGVILTHKSLTTSVAQQVDGENPNLYLTTEDVLLCVLPLF 254
Query: 567 HIFSLNSVLLCALRAGSGVLLMPKFEIGSLLELIQKHRVSVAMVVPPLVLALAKNPKVAE 388
HIFSLNSVLLCALRAGS VLLM KFEIG+LLELIQ+HRVSVAMVVPPLVLALAKNP VA+
Sbjct: 255 HIFSLNSVLLCALRAGSAVLLMQKFEIGTLLELIQRHRVSVAMVVPPLVLALAKNPMVAD 314
Query: 387 FDLSSIRLVLSGAAPLGKELEEALRSRVPQAVLGQGYGMTEAGPVLSMSLGFAKQPFPTK 208
FDLSSIRLVLSGAAPLGKELEEALR+R+PQAVLGQGYGMTEAGPVLSM LGFAKQPF TK
Sbjct: 315 FDLSSIRLVLSGAAPLGKELEEALRNRMPQAVLGQGYGMTEAGPVLSMCLGFAKQPFQTK 374
Query: 207 SGSCGTVVRNAELKVLDPETGCSLGYNQPGEICIRGQQIMKGYLNDEKATATTIDAEGWL 28
SGSCGTVVRNAELKV+DPETG SLGYNQPGEICIRGQQIMKGYLNDE ATA+TID+EGWL
Sbjct: 375 SGSCGTVVRNAELKVVDPETGRSLGYNQPGEICIRGQQIMKGYLNDEAATASTIDSEGWL 434
Query: 27 HTGDVGYID 1
HTGDVGY+D
Sbjct: 435 HTGDVGYVD 443
>pir||T08075 4-coumarate-CoA ligase (EC 6.2.1.12) (clone 4CL2) [validated] -
quaking aspen gi|3258637|gb|AAC24504.1| 4-coumarate:CoA
ligase [Populus tremuloides]
Length = 570
Score = 476 bits (1225), Expect = e-133
Identities = 240/309 (77%), Positives = 266/309 (85%)
Frame = -2
Query: 927 KLVITQAMYVDKLRPLEDVNGNDYPKLGEDFKVVTVDEQPKDCLDFSVISEGKEDDLPEV 748
KL+ITQ+ YV+KL + N P GEDF V+T+D+ P++CL F+V+ E E ++P V
Sbjct: 146 KLIITQSQYVNKLGDSDCHENNQKP--GEDFIVITIDDPPENCLHFNVLVEASESEMPTV 203
Query: 747 EINPEDAVALPFSSGTTGLPKGVVLTHKSLTTSVGQQVDGENPNLSLGTEDVLLCVLPLF 568
I P+D VALPFSSGTTGLPKGV+LTHKSL TSV QQVDGE PNL L +DV+LCVLPLF
Sbjct: 204 SILPDDPVALPFSSGTTGLPKGVILTHKSLITSVAQQVDGEIPNLYLKQDDVVLCVLPLF 263
Query: 567 HIFSLNSVLLCALRAGSGVLLMPKFEIGSLLELIQKHRVSVAMVVPPLVLALAKNPKVAE 388
HIFSLNSVLLC+LRAGS VLLM KFEIGSLLELIQKH VSVA VVPPLVLALAKNP A
Sbjct: 264 HIFSLNSVLLCSLRAGSAVLLMQKFEIGSLLELIQKHNVSVAAVVPPLVLALAKNPLEAN 323
Query: 387 FDLSSIRLVLSGAAPLGKELEEALRSRVPQAVLGQGYGMTEAGPVLSMSLGFAKQPFPTK 208
FDLSSIR+VLSGAAPLGKELE+ALRSRVPQA+LGQGYGMTEAGPVLSM L F+KQPFPTK
Sbjct: 324 FDLSSIRVVLSGAAPLGKELEDALRSRVPQAILGQGYGMTEAGPVLSMCLAFSKQPFPTK 383
Query: 207 SGSCGTVVRNAELKVLDPETGCSLGYNQPGEICIRGQQIMKGYLNDEKATATTIDAEGWL 28
SGSCGTVVRNAELKV+DPETG SLGYNQPGEICIRG QIMKGYLND +ATA TID EGWL
Sbjct: 384 SGSCGTVVRNAELKVIDPETGRSLGYNQPGEICIRGSQIMKGYLNDAEATANTIDVEGWL 443
Query: 27 HTGDVGYID 1
HTGD+GY+D
Sbjct: 444 HTGDIGYVD 452
>gb|AAK58909.1| 4-coumarate:CoA ligase 4 [Populus balsamifera subsp. trichocarpa x
Populus deltoides]
Length = 579
Score = 473 bits (1217), Expect = e-132
Identities = 238/309 (77%), Positives = 266/309 (86%)
Frame = -2
Query: 927 KLVITQAMYVDKLRPLEDVNGNDYPKLGEDFKVVTVDEQPKDCLDFSVISEGKEDDLPEV 748
KL+ITQ+ +V+KLR + N P+ EDF V+T+D+ P++CL F+V+ E E ++P V
Sbjct: 146 KLIITQSQHVNKLRDSDYHENNQKPE--EDFIVITIDDPPENCLHFNVLVEANESEMPTV 203
Query: 747 EINPEDAVALPFSSGTTGLPKGVVLTHKSLTTSVGQQVDGENPNLSLGTEDVLLCVLPLF 568
I+P+D VALPFSSGTTGLPKGV+LTHKSL TSV QQVDGE PNL L +DV+LCVLPLF
Sbjct: 204 SIHPDDPVALPFSSGTTGLPKGVILTHKSLITSVAQQVDGEIPNLYLKQDDVVLCVLPLF 263
Query: 567 HIFSLNSVLLCALRAGSGVLLMPKFEIGSLLELIQKHRVSVAMVVPPLVLALAKNPKVAE 388
HIFSLNSVLLC+LRAGS VLLM KFEIGSLLELIQKH VSVA VVPPLVLALAKNP VA
Sbjct: 264 HIFSLNSVLLCSLRAGSAVLLMQKFEIGSLLELIQKHNVSVAAVVPPLVLALAKNPMVAN 323
Query: 387 FDLSSIRLVLSGAAPLGKELEEALRSRVPQAVLGQGYGMTEAGPVLSMSLGFAKQPFPTK 208
FDLSSIR+VLSGAAPLGKELEEALRSRVPQA+LGQGYGMTEAGPVLSM L F+KQP PTK
Sbjct: 324 FDLSSIRVVLSGAAPLGKELEEALRSRVPQAILGQGYGMTEAGPVLSMCLAFSKQPLPTK 383
Query: 207 SGSCGTVVRNAELKVLDPETGCSLGYNQPGEICIRGQQIMKGYLNDEKATATTIDAEGWL 28
SGSCGTVVRNAELKV+DPETG SLG NQPGEICIRG QIMKGYLND +ATA ID EGWL
Sbjct: 384 SGSCGTVVRNAELKVIDPETGSSLGRNQPGEICIRGSQIMKGYLNDAEATANIIDVEGWL 443
Query: 27 HTGDVGYID 1
HTGD+GY+D
Sbjct: 444 HTGDIGYVD 452
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 909,991,729
Number of Sequences: 1393205
Number of extensions: 24197003
Number of successful extensions: 216426
Number of sequences better than 10.0: 5567
Number of HSP's better than 10.0 without gapping: 113442
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 171004
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 51305130920
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)