Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011088A_C01 KMC011088A_c01
(529 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
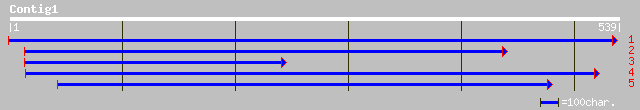
ref|NP_198322.1| putative protein; protein id: At5g33370.1, supp... 135 3e-31
ref|NP_187079.1| putative GDSL-motif lipase/acylhydrolase; prote... 131 6e-30
dbj|BAC56011.1| P0705A05.30 [Oryza sativa (japonica cultivar-gro... 128 5e-29
ref|NP_194607.1| Proline-rich APG - like protein; protein id: At... 125 3e-28
ref|NP_197344.1| putative protein; protein id: At5g18430.1 [Arab... 123 2e-27
>ref|NP_198322.1| putative protein; protein id: At5g33370.1, supported by cDNA:
34612. [Arabidopsis thaliana] gi|21592967|gb|AAM64916.1|
putative GDSL-motif lipase/acylhydrolase [Arabidopsis
thaliana] gi|28393449|gb|AAO42146.1| unknown protein
[Arabidopsis thaliana] gi|28827340|gb|AAO50514.1|
unknown protein [Arabidopsis thaliana]
Length = 366
Score = 135 bits (340), Expect = 3e-31
Identities = 56/77 (72%), Positives = 70/77 (90%)
Frame = -2
Query: 528 AFGFVTSKVACCGQGPYNGLGLCTTASNLCPDRDVYAFWDPFHPSERANSFIVQQIMSGN 349
A+GFVTSKVACCGQGPYNG+GLCT SNLCP+RD++AFWDPFHPSE+A+ I QQI++G+
Sbjct: 288 AYGFVTSKVACCGQGPYNGIGLCTPLSNLCPNRDLFAFWDPFHPSEKASRIIAQQILNGS 347
Query: 348 TEYMYPMNLSTVLAIDA 298
EYM+PMNLST+L +D+
Sbjct: 348 PEYMHPMNLSTILTVDS 364
>ref|NP_187079.1| putative GDSL-motif lipase/acylhydrolase; protein id: At3g04290.1,
supported by cDNA: 1323. [Arabidopsis thaliana]
gi|6721157|gb|AAF26785.1|AC016829_9 putative GDSL-motif
lipase/acylhydrolase [Arabidopsis thaliana]
gi|21537340|gb|AAM61681.1| putative GDSL-motif
lipase/acylhydrolase [Arabidopsis thaliana]
Length = 366
Score = 131 bits (329), Expect = 6e-30
Identities = 54/77 (70%), Positives = 69/77 (89%)
Frame = -2
Query: 525 FGFVTSKVACCGQGPYNGLGLCTTASNLCPDRDVYAFWDPFHPSERANSFIVQQIMSGNT 346
FGFVTSKVACCGQGPYNG+GLCT SNLCP+RD+YAFWD FHP+E+AN IV QI++G++
Sbjct: 288 FGFVTSKVACCGQGPYNGIGLCTPVSNLCPNRDLYAFWDAFHPTEKANRIIVNQILTGSS 347
Query: 345 EYMYPMNLSTVLAIDAA 295
+YM+PMNLST + +D++
Sbjct: 348 KYMHPMNLSTAMLLDSS 364
>dbj|BAC56011.1| P0705A05.30 [Oryza sativa (japonica cultivar-group)]
Length = 387
Score = 128 bits (321), Expect = 5e-29
Identities = 55/78 (70%), Positives = 64/78 (81%)
Frame = -2
Query: 528 AFGFVTSKVACCGQGPYNGLGLCTTASNLCPDRDVYAFWDPFHPSERANSFIVQQIMSGN 349
A+GF TSKVACCGQGPYNG+GLCT S LCPDR +Y FWD FHP+ERAN IV Q MS +
Sbjct: 304 AYGFATSKVACCGQGPYNGVGLCTALSTLCPDRSLYVFWDNFHPTERANRIIVSQFMSAS 363
Query: 348 TEYMYPMNLSTVLAIDAA 295
+YM+P NLST+LA+DAA
Sbjct: 364 PDYMHPFNLSTILAMDAA 381
>ref|NP_194607.1| Proline-rich APG - like protein; protein id: At4g28780.1, supported
by cDNA: 324. [Arabidopsis thaliana]
gi|7488228|pir||T04521 proline-rich protein APG homolog
F16A16.110 - Arabidopsis thaliana
gi|4218120|emb|CAA22974.1| Proline-rich APG-like protein
[Arabidopsis thaliana] gi|7269733|emb|CAB81466.1|
Proline-rich APG-like protein [Arabidopsis thaliana]
gi|21592773|gb|AAM64722.1| Proline-rich APG-like protein
[Arabidopsis thaliana] gi|27754717|gb|AAO22802.1|
putative proline-rich APG protein [Arabidopsis thaliana]
gi|28394103|gb|AAO42459.1| putative proline-rich APG
protein [Arabidopsis thaliana]
Length = 367
Score = 125 bits (314), Expect = 3e-28
Identities = 54/76 (71%), Positives = 64/76 (84%)
Frame = -2
Query: 525 FGFVTSKVACCGQGPYNGLGLCTTASNLCPDRDVYAFWDPFHPSERANSFIVQQIMSGNT 346
FGFVTSKVACCGQG YNG G+CT S LC DR+ YAFWDPFHP+E+A IVQQIM+G+
Sbjct: 290 FGFVTSKVACCGQGAYNGQGVCTPLSTLCSDRNAYAFWDPFHPTEKATRLIVQQIMTGSV 349
Query: 345 EYMYPMNLSTVLAIDA 298
EYM PMNLST++A+D+
Sbjct: 350 EYMNPMNLSTIMALDS 365
>ref|NP_197344.1| putative protein; protein id: At5g18430.1 [Arabidopsis thaliana]
Length = 349
Score = 123 bits (308), Expect = 2e-27
Identities = 51/74 (68%), Positives = 64/74 (85%)
Frame = -2
Query: 525 FGFVTSKVACCGQGPYNGLGLCTTASNLCPDRDVYAFWDPFHPSERANSFIVQQIMSGNT 346
+GFVTSKVACCGQGPYNG+GLCT SNLCP+R++Y FWD FHP+E+AN IV+ I++G T
Sbjct: 276 YGFVTSKVACCGQGPYNGMGLCTVLSNLCPNRELYVFWDAFHPTEKANRMIVRHILTGTT 335
Query: 345 EYMYPMNLSTVLAI 304
+YM PMNLS+ LA+
Sbjct: 336 KYMNPMNLSSALAL 349
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 436,425,363
Number of Sequences: 1393205
Number of extensions: 8829049
Number of successful extensions: 18719
Number of sequences better than 10.0: 202
Number of HSP's better than 10.0 without gapping: 17995
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18656
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17308240320
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)