Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC011066A_C01 KMC011066A_c01
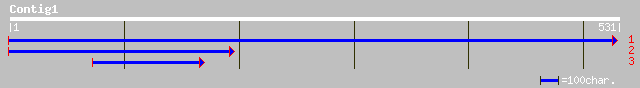
(531 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF68118.1|AC010793_13 F20B17.16 [Arabidopsis thaliana] 98 5e-20
ref|NP_178091.2| unknown protein; protein id: At1g79730.1, suppo... 98 5e-20
gb|AAM10397.1| At1g79730/F19K16_29 [Arabidopsis thaliana] gi|273... 98 5e-20
ref|NP_800472.1| putative formate dehydrogenase oxidoreductase p... 33 2.7
emb|CAB91734.2| hypothetical protein [Neurospora crassa] gi|2892... 33 2.7
>gb|AAF68118.1|AC010793_13 F20B17.16 [Arabidopsis thaliana]
Length = 593
Score = 98.2 bits (243), Expect = 5e-20
Identities = 54/99 (54%), Positives = 66/99 (66%), Gaps = 1/99 (1%)
Frame = -2
Query: 521 ARYVPLPTKLVLRKKRATEGRSGDEVEPFPIPSKVTVRRRSSVAAIERKDSGVYTSSKGN 342
A Y+PLP +L LRKKRA EGRS DE+E FP+PS+VTVRRRS+V+ IE KDSGVY+S G
Sbjct: 495 ASYLPLPMRLNLRKKRAREGRSSDEIEHFPVPSRVTVRRRSTVSVIEHKDSGVYSSRVGA 554
Query: 341 SSRRGDLEMDDDALEHQHRVAAHQD-NYQSSGAEDDMSD 228
SS + D+ L + QD N S G EDD S+
Sbjct: 555 SSSKMRRLEDEGGLGRSWKHEPEQDANQYSDGNEDDYSE 593
>ref|NP_178091.2| unknown protein; protein id: At1g79730.1, supported by cDNA:
gi_20147373 [Arabidopsis thaliana]
Length = 589
Score = 98.2 bits (243), Expect = 5e-20
Identities = 54/99 (54%), Positives = 66/99 (66%), Gaps = 1/99 (1%)
Frame = -2
Query: 521 ARYVPLPTKLVLRKKRATEGRSGDEVEPFPIPSKVTVRRRSSVAAIERKDSGVYTSSKGN 342
A Y+PLP +L LRKKRA EGRS DE+E FP+PS+VTVRRRS+V+ IE KDSGVY+S G
Sbjct: 491 ASYLPLPMRLNLRKKRAREGRSSDEIEHFPVPSRVTVRRRSTVSVIEHKDSGVYSSRVGA 550
Query: 341 SSRRGDLEMDDDALEHQHRVAAHQD-NYQSSGAEDDMSD 228
SS + D+ L + QD N S G EDD S+
Sbjct: 551 SSSKMRRLEDEGGLGRSWKHEPEQDANQYSDGNEDDYSE 589
>gb|AAM10397.1| At1g79730/F19K16_29 [Arabidopsis thaliana]
gi|27363416|gb|AAO11627.1| At1g79730/F19K16_29
[Arabidopsis thaliana]
Length = 589
Score = 98.2 bits (243), Expect = 5e-20
Identities = 54/99 (54%), Positives = 66/99 (66%), Gaps = 1/99 (1%)
Frame = -2
Query: 521 ARYVPLPTKLVLRKKRATEGRSGDEVEPFPIPSKVTVRRRSSVAAIERKDSGVYTSSKGN 342
A Y+PLP +L LRKKRA EGRS DE+E FP+PS+VTVRRRS+V+ IE KDSGVY+S G
Sbjct: 491 ASYLPLPMRLNLRKKRAREGRSSDEIEHFPVPSRVTVRRRSTVSVIEHKDSGVYSSRVGA 550
Query: 341 SSRRGDLEMDDDALEHQHRVAAHQD-NYQSSGAEDDMSD 228
SS + D+ L + QD N S G EDD S+
Sbjct: 551 SSSKMRRLEDEGGLGRSWKHEPEQDANQYSDGNEDDYSE 589
>ref|NP_800472.1| putative formate dehydrogenase oxidoreductase protein [Vibrio
parahaemolyticus RIMD 2210633]
gi|28809263|dbj|BAC62305.1| putative formate
dehydrogenase oxidoreductase protein [Vibrio
parahaemolyticus]
Length = 276
Score = 32.7 bits (73), Expect = 2.7
Identities = 17/32 (53%), Positives = 22/32 (68%), Gaps = 1/32 (3%)
Frame = -1
Query: 303 SRTPT*SC-STPGQLSVEWSRR*HV*LIH*PQ 211
+R PT C S P L+VEW+RR H+ LIH P+
Sbjct: 234 ARIPTLVCLSAPTALTVEWARRNHLNLIHLPK 265
>emb|CAB91734.2| hypothetical protein [Neurospora crassa] gi|28925318|gb|EAA34396.1|
predicted protein [Neurospora crassa]
Length = 659
Score = 32.7 bits (73), Expect = 2.7
Identities = 31/103 (30%), Positives = 46/103 (44%), Gaps = 22/103 (21%)
Frame = -2
Query: 485 RKKR----ATEGRSGDEVEPFPIPSKVT---------------VRRRSSVAAIERK---D 372
RK+R A E DEV+P P P+K VR R VA+IE + +
Sbjct: 135 RKRRRPTPAAEESVEDEVQPDPEPTKRPRGRPSKDNGKATAKGVRGRKEVASIEEEPAQE 194
Query: 371 SGVYTSSKGNSSRRGDLEMDDDALEHQHRVAAHQDNYQSSGAE 243
V S++ + + + E++ +AHQDN +SS AE
Sbjct: 195 GDVRRSTRSTNPSQHSSKTTQRGKENRRDPSAHQDNGRSSLAE 237
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 437,463,300
Number of Sequences: 1393205
Number of extensions: 9023948
Number of successful extensions: 23714
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 22964
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23698
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17596710992
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)