Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010956A_C01 KMC010956A_c01
(642 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
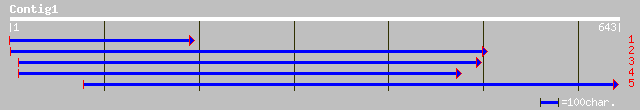
ref|NP_194473.1| COP1-interacting protein 7 (CIP7); protein id: ... 57 3e-07
ref|NP_593746.1| hypothetical protein [Schizosaccharomyces pombe... 32 5.5
ref|NP_595849.1| hypothetical protein; possible coiled-coil regi... 32 7.2
pdb|1A0T|P Chain P, Sucrose-Specific Porin, With Bound Sucrose M... 32 9.4
ref|ZP_00128127.1| hypothetical protein [Pseudomonas syringae pv... 32 9.4
>ref|NP_194473.1| COP1-interacting protein 7 (CIP7); protein id: At4g27430.1, supported
by cDNA: gi_3327867 [Arabidopsis thaliana]
gi|7484877|pir||T08935 COP1-interacting protein 7 -
Arabidopsis thaliana gi|3327868|dbj|BAA31738.1|
COP1-Interacting Protein 7 (CIP7) [Arabidopsis thaliana]
gi|4972068|emb|CAB43875.1| COP1-interacting protein 7
(CIP7) [Arabidopsis thaliana] gi|7269597|emb|CAB81393.1|
COP1-interacting protein 7 (CIP7) [Arabidopsis thaliana]
Length = 1058
Score = 56.6 bits (135), Expect = 3e-07
Identities = 33/61 (54%), Positives = 40/61 (65%), Gaps = 1/61 (1%)
Frame = -2
Query: 635 IDESNAVPPKVSEIQISTPPPRPSNQVMSESIHSRKKWNNDEDSLKA-AKGFRKLLFFGR 459
++E P VSEI ISTPP +E+ HSRKKWN++E S KA AK FRKLL FGR
Sbjct: 1004 MEEKYYFSPAVSEIDISTPPA-------TEADHSRKKWNSEETSPKATAKVFRKLLMFGR 1056
Query: 458 R 456
+
Sbjct: 1057 K 1057
>ref|NP_593746.1| hypothetical protein [Schizosaccharomyces pombe]
gi|1723464|sp|Q10302|YD49_SCHPO Hypothetical protein
C22H10.09 in chromosome I gi|7490960|pir||T38212
hypothetical protein SPAC22H10.09 - fission yeast
(Schizosaccharomyces pombe) gi|1204190|emb|CAA93610.1|
hypothetical protein [Schizosaccharomyces pombe]
Length = 646
Score = 32.3 bits (72), Expect = 5.5
Identities = 11/48 (22%), Positives = 28/48 (57%)
Frame = -1
Query: 477 ASLLWKKELKHNGGF*FFANGFTIIFSKCSEKLHPWFWLFNISTKIYF 334
++L W+ +N + + I++KC + LHP++ +F +++ +Y+
Sbjct: 66 STLAWRDYENYNDDDLLYQRELSEIYNKCLKALHPFYPVFEVNSNLYY 113
>ref|NP_595849.1| hypothetical protein; possible coiled-coil region
[Schizosaccharomyces pombe] gi|7491444|pir||T39754
hypothetical protein SPBC18E5.03c - fission yeast
(Schizosaccharomyces pombe) gi|4107322|emb|CAA22663.1|
hypothetical protein; possible coiled-coil region
[Schizosaccharomyces pombe]
Length = 277
Score = 32.0 bits (71), Expect = 7.2
Identities = 22/89 (24%), Positives = 45/89 (49%), Gaps = 3/89 (3%)
Frame = -2
Query: 623 NAVPPKVSEIQISTPPPRPSNQVMSESIHSRKKWNNDEDSLKAAKGFR--KLLFFGRRS* 450
NA+ +S+++ + + V+ E +H KK+++D SLK++ G R + RR
Sbjct: 92 NAIDNNISDLKKNLHSNKKLEAVLKEELHQIKKFSSDLQSLKSSMGERQKQQAAMSRRGK 151
Query: 449 STMVDFDFLQMASQ*SSQS-VLRSFIHGF 366
+++ +Q + +S L +F+ GF
Sbjct: 152 KSIIHSAIIQEKERYEKESQELFTFLDGF 180
>pdb|1A0T|P Chain P, Sucrose-Specific Porin, With Bound Sucrose Molecules
gi|2981660|pdb|1A0T|Q Chain Q, Sucrose-Specific Porin,
With Bound Sucrose Molecules gi|2981661|pdb|1A0T|R Chain
R, Sucrose-Specific Porin, With Bound Sucrose Molecules
gi|3319010|pdb|1A0S|P Chain P, Sucrose-Specific Porin
gi|3319011|pdb|1A0S|Q Chain Q, Sucrose-Specific Porin
gi|3319012|pdb|1A0S|R Chain R, Sucrose-Specific Porin
Length = 413
Score = 31.6 bits (70), Expect = 9.4
Identities = 11/21 (52%), Positives = 16/21 (75%)
Frame = +1
Query: 523 HFFLEWIDSDITWLLGLGGGV 585
+F + WIDSD+ +L G GGG+
Sbjct: 122 NFDIHWIDSDVVFLAGTGGGI 142
>ref|ZP_00128127.1| hypothetical protein [Pseudomonas syringae pv. syringae B728a]
Length = 459
Score = 31.6 bits (70), Expect = 9.4
Identities = 17/45 (37%), Positives = 27/45 (59%), Gaps = 13/45 (28%)
Frame = +1
Query: 490 LAAFK-----ESSSLF--------HFFLEWIDSDITWLLGLGGGV 585
+AAFK E+S+L+ +F + W+DSD+ +L G GGG+
Sbjct: 144 IAAFKGNSVFENSTLWAGKRFDRDNFDIHWLDSDVVFLAGTGGGI 188
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 577,477,787
Number of Sequences: 1393205
Number of extensions: 12551439
Number of successful extensions: 30456
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 29360
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30433
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27007650415
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)