Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010934A_C01 KMC010934A_c01
(885 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
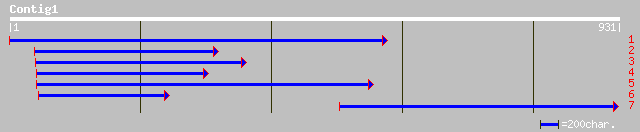
ref|NP_564014.1| ubiquitin-specific protease 15 (UBP15); protein... 71 2e-11
gb|AAG42756.1|AF302665_1 ubiquitin-specific protease 15 [Arabido... 71 2e-11
pir||H86306 F20D23.20 protein - Arabidopsis thaliana gi|5734755|... 71 2e-11
ref|NP_043464.1| major capsid protein [Leishmania RNA virus 2-1]... 36 0.68
gb|EAA11154.1| agCP5532 [Anopheles gambiae str. PEST] 36 0.88
>ref|NP_564014.1| ubiquitin-specific protease 15 (UBP15); protein id: At1g17110.1,
supported by cDNA: gi_11993474 [Arabidopsis thaliana]
Length = 924
Score = 70.9 bits (172), Expect = 2e-11
Identities = 41/91 (45%), Positives = 58/91 (63%), Gaps = 14/91 (15%)
Frame = -2
Query: 776 YAEKVRHEFSDATSSDWSLFTSSDEASFTTESTRDSFSTVDYGDNMDPIS-----AIFNY 612
+ + H+ ++++SS+WSLFTSSDEASFTTESTRDSFST+DY D + AIFN
Sbjct: 778 FKPRADHKNTESSSSEWSLFTSSDEASFTTESTRDSFSTIDYTDVCHVVDSSSPFAIFNN 837
Query: 611 -------TPENSL--MKFSHSRPLTRFIPEK 546
+P N++ FS ++P TR+ E+
Sbjct: 838 VYHNVEPSPHNTVACRMFSGTKPETRYFVEQ 868
>gb|AAG42756.1|AF302665_1 ubiquitin-specific protease 15 [Arabidopsis thaliana]
Length = 924
Score = 70.9 bits (172), Expect = 2e-11
Identities = 41/91 (45%), Positives = 58/91 (63%), Gaps = 14/91 (15%)
Frame = -2
Query: 776 YAEKVRHEFSDATSSDWSLFTSSDEASFTTESTRDSFSTVDYGDNMDPIS-----AIFNY 612
+ + H+ ++++SS+WSLFTSSDEASFTTESTRDSFST+DY D + AIFN
Sbjct: 778 FKPRADHKNTESSSSEWSLFTSSDEASFTTESTRDSFSTIDYTDVCHVVDSSSPFAIFNN 837
Query: 611 -------TPENSL--MKFSHSRPLTRFIPEK 546
+P N++ FS ++P TR+ E+
Sbjct: 838 VYHNVEPSPHNTVACRMFSGTKPETRYFVEQ 868
>pir||H86306 F20D23.20 protein - Arabidopsis thaliana
gi|5734755|gb|AAD50020.1|AC007651_15 Unknown protein
[Arabidopsis thaliana]
Length = 891
Score = 70.9 bits (172), Expect = 2e-11
Identities = 41/91 (45%), Positives = 58/91 (63%), Gaps = 14/91 (15%)
Frame = -2
Query: 776 YAEKVRHEFSDATSSDWSLFTSSDEASFTTESTRDSFSTVDYGDNMDPIS-----AIFNY 612
+ + H+ ++++SS+WSLFTSSDEASFTTESTRDSFST+DY D + AIFN
Sbjct: 745 FKPRADHKNTESSSSEWSLFTSSDEASFTTESTRDSFSTIDYTDVCHVVDSSSPFAIFNN 804
Query: 611 -------TPENSL--MKFSHSRPLTRFIPEK 546
+P N++ FS ++P TR+ E+
Sbjct: 805 VYHNVEPSPHNTVACRMFSGTKPETRYFVEQ 835
>ref|NP_043464.1| major capsid protein [Leishmania RNA virus 2-1]
gi|1045466|gb|AAB50030.1| major capsid protein
Length = 714
Score = 36.2 bits (82), Expect = 0.68
Identities = 22/119 (18%), Positives = 50/119 (41%), Gaps = 2/119 (1%)
Frame = -2
Query: 863 PEIATSLIDTSNG--FLTSNTNRNIHPVTQTYAEKVRHEFSDATSSDWSLFTSSDEASFT 690
P++ TS+ + + G ++ +T+ ++ Y VR + T+ W L +AS
Sbjct: 6 PQMNTSVFNLTRGRRVVSGDTSERVYTAVLNYCSTVRQSYQATTNVHWRLKQRGKKASHV 65
Query: 689 TESTRDSFSTVDYGDNMDPISAIFNYTPENSLMKFSHSRPLTRFIPEKGHV*QFRELIN 513
+ + ++ DY DN+ + ++K+ T + + + FR L++
Sbjct: 66 SRAHAGISASFDYADNL-----------KQEILKYHSRHVCTEIVVGRDYRFDFRSLLH 113
>gb|EAA11154.1| agCP5532 [Anopheles gambiae str. PEST]
Length = 158
Score = 35.8 bits (81), Expect = 0.88
Identities = 26/72 (36%), Positives = 32/72 (44%), Gaps = 13/72 (18%)
Frame = -2
Query: 818 TSNTNRNIHPVTQTYAEK-VRHEFSDATSS------------DWSLFTSSDEASFTTEST 678
TSN+ PVT A K V + +T S +WS+F SSD S T + T
Sbjct: 36 TSNSPAPDKPVTMAEASKGVPQQSVPSTDSLEESDVDVGSEDEWSIFDSSDPYSETDDDT 95
Query: 677 RDSFSTVDYGDN 642
DS DYG N
Sbjct: 96 EDSTDDEDYGSN 107
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 702,004,694
Number of Sequences: 1393205
Number of extensions: 14162305
Number of successful extensions: 31015
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 30045
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30981
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 47939536764
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)