Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010921A_C01 KMC010921A_c01
(671 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
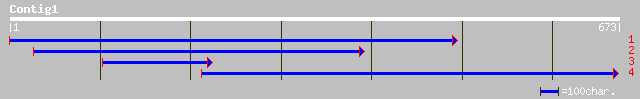
ref|NP_567601.1| subtilisin-like serine protease; protein id: At... 176 3e-43
ref|NP_568634.1| subtilisin-like serine protease; protein id: At... 174 1e-42
ref|NP_174348.1| putative serine proteinase; protein id: At1g306... 172 5e-42
dbj|BAB56061.1| putative serine proteinase [Oryza sativa (japoni... 156 3e-37
gb|AAF13299.1|AF181496_1 meiotic serine proteinase [Lycopersicon... 84 2e-15
>ref|NP_567601.1| subtilisin-like serine protease; protein id: At4g20430.1 [Arabidopsis
thaliana] gi|7435683|pir||T10585 serine proteinase
homolog F9F13.80 - Arabidopsis thaliana
gi|5262166|emb|CAB45809.1| putative serine proteinase
[Arabidopsis thaliana] gi|7268839|emb|CAB79043.1|
putative serine proteinase [Arabidopsis thaliana]
Length = 856
Score = 176 bits (445), Expect = 3e-43
Identities = 87/138 (63%), Positives = 108/138 (78%)
Frame = -1
Query: 671 SSYDDYMSFLCGINGSAPVVLNYTGQNCGSYNSTVLGPDLNLPSITLSKLNPSRIVQRTV 492
+S++DYMSFLCGINGSAPVV NYTG NC N+T+ G DLNLPSIT+SKLN +R VQR +
Sbjct: 717 TSFEDYMSFLCGINGSAPVVFNYTGTNCLRNNATISGSDLNLPSITVSKLNNTRTVQRLM 776
Query: 491 LNIAGNETYSVGWDAPSGVSVKVTPSHFTIAHGERKVLSVLLNATLNSTVASFGRIGLFG 312
NIAGNETY+V P V + V+P+ F+IA GE K+LSV+L A NS+++SFG I L G
Sbjct: 777 TNIAGNETYTVSLITPFDVLINVSPTQFSIASGETKLLSVILTAKRNSSISSFGGIKLLG 836
Query: 311 NQGHVVNIPVSVTVKISN 258
N GH+V IPVSVTVKI++
Sbjct: 837 NAGHIVRIPVSVTVKIAS 854
>ref|NP_568634.1| subtilisin-like serine protease; protein id: At5g44530.1 [Arabidopsis
thaliana] gi|9758706|dbj|BAB09160.1| serine proteinase
[Arabidopsis thaliana]
Length = 840
Score = 174 bits (441), Expect = 1e-42
Identities = 85/139 (61%), Positives = 110/139 (78%)
Frame = -1
Query: 671 SSYDDYMSFLCGINGSAPVVLNYTGQNCGSYNSTVLGPDLNLPSITLSKLNPSRIVQRTV 492
+S++DY+SFLCGINGS VV NYTG C + N+ V G DLNLPSIT+S L+ ++ QR++
Sbjct: 702 TSFEDYISFLCGINGSDTVVFNYTGFRCPANNTPVSGFDLNLPSITVSTLSGTQTFQRSM 761
Query: 491 LNIAGNETYSVGWDAPSGVSVKVTPSHFTIAHGERKVLSVLLNATLNSTVASFGRIGLFG 312
NIAGNETY+VGW P GVS+KV+P+ F+IA GE +VLSV L T NS+ +SFGRIGLFG
Sbjct: 762 RNIAGNETYNVGWSPPYGVSMKVSPTQFSIAMGENQVLSVTLTVTKNSSSSSFGRIGLFG 821
Query: 311 NQGHVVNIPVSVTVKISNN 255
N GH+VNIPV+V KI+++
Sbjct: 822 NTGHIVNIPVTVIAKIASS 840
>ref|NP_174348.1| putative serine proteinase; protein id: At1g30600.1, supported by
cDNA: gi_18175991, supported by cDNA: gi_20465292
[Arabidopsis thaliana] gi|25289831|pir||C86431 T5I8.5
protein - Arabidopsis thaliana
gi|4587516|gb|AAD25747.1|AC007060_5 Strong similarity to
gb|U80583 proteinase TMP from Lycopersicon esculentum and
is a member of the PF|00082 subtilase family.
[Arabidopsis thaliana] gi|18175992|gb|AAL59964.1|
putative serine proteinase [Arabidopsis thaliana]
gi|20465293|gb|AAM20050.1| putative serine proteinase
[Arabidopsis thaliana]
Length = 832
Score = 172 bits (435), Expect = 5e-42
Identities = 85/138 (61%), Positives = 109/138 (78%), Gaps = 3/138 (2%)
Frame = -1
Query: 665 YDDYMSFLCGINGSAPVVLNYTGQNCGSYNSTVLGPDLNLPSITLSKLNPSRIVQRTVLN 486
Y++YM FLCGINGS+PVVLNYTG++C SYNS++ DLNLPS+T++KL +R V R V N
Sbjct: 694 YNEYMKFLCGINGSSPVVLNYTGESCSSYNSSLAASDLNLPSVTIAKLVGTRAVLRWVTN 753
Query: 485 IAG---NETYSVGWDAPSGVSVKVTPSHFTIAHGERKVLSVLLNATLNSTVASFGRIGLF 315
IA NETY VGW AP VSVKV+P+ FTI +G+ +VLS++ A N ++ASFGRIGLF
Sbjct: 754 IATTATNETYIVGWMAPDSVSVKVSPAKFTIGNGQTRVLSLVFRAMKNVSMASFGRIGLF 813
Query: 314 GNQGHVVNIPVSVTVKIS 261
G++GHVVNIPV+V KI+
Sbjct: 814 GDRGHVVNIPVAVIYKIA 831
>dbj|BAB56061.1| putative serine proteinase [Oryza sativa (japonica cultivar-group)]
gi|21743316|dbj|BAC03311.1| putative serine proteinase
[Oryza sativa (japonica cultivar-group)]
Length = 849
Score = 156 bits (394), Expect = 3e-37
Identities = 75/136 (55%), Positives = 101/136 (74%)
Frame = -1
Query: 671 SSYDDYMSFLCGINGSAPVVLNYTGQNCGSYNSTVLGPDLNLPSITLSKLNPSRIVQRTV 492
SSYDD+ SFLCGINGSAPVV+NYTG +C S S + G DLNLPSIT++ LN SR + RTV
Sbjct: 712 SSYDDFFSFLCGINGSAPVVMNYTGNSCSS--SAMTGADLNLPSITIAVLNQSRTITRTV 769
Query: 491 LNIAGNETYSVGWDAPSGVSVKVTPSHFTIAHGERKVLSVLLNATLNSTVASFGRIGLFG 312
N+A +E Y+V + AP GV+V +P+ F I G+R+ ++ ++NAT+N T ASFG +G +G
Sbjct: 770 TNVASDERYTVSYSAPYGVAVSASPAQFFIPSGQRQQVTFVVNATMNGTSASFGSVGFYG 829
Query: 311 NQGHVVNIPVSVTVKI 264
++GH V IP SV K+
Sbjct: 830 DKGHRVMIPFSVISKV 845
>gb|AAF13299.1|AF181496_1 meiotic serine proteinase [Lycopersicon esculentum]
Length = 809
Score = 84.0 bits (206), Expect = 2e-15
Identities = 56/136 (41%), Positives = 72/136 (52%), Gaps = 2/136 (1%)
Frame = -1
Query: 671 SSYDDYMSFLCGINGSAPV-VLNYTGQNCGSYNSTVLGPDLNLPSITLSKLNPSRIVQRT 495
+S+ Y+ FLC + G + V G C S DLN PS+T+S L SR V R
Sbjct: 669 ASFKHYVLFLCSVPGVDEMSVRRAVGVGCPSKKKAWCS-DLNTPSVTISNLVGSRNVIRR 727
Query: 494 VLNIAG-NETYSVGWDAPSGVSVKVTPSHFTIAHGERKVLSVLLNATLNSTVASFGRIGL 318
V N+AG +ETY V P GVSV V P F I K ++ +LNAT + SFG I
Sbjct: 728 VTNVAGVDETYQVIVQEPLGVSVTVRPRVFNIIAKASKHITFVLNATQTTNTYSFGEIVF 787
Query: 317 FGNQGHVVNIPVSVTV 270
GNQ H V +P++V V
Sbjct: 788 QGNQNHTVRVPLAVFV 803
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 546,735,171
Number of Sequences: 1393205
Number of extensions: 11214524
Number of successful extensions: 23673
Number of sequences better than 10.0: 152
Number of HSP's better than 10.0 without gapping: 22930
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23627
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29421376608
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)