Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
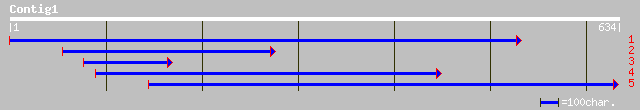
Query= KMC010920A_C01 KMC010920A_c01
(633 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_564567.1| expressed protein; protein id: At1g50380.1, sup... 121 7e-27
ref|NP_799677.1| putative protease [Vibrio parahaemolyticus RIMD... 86 5e-16
gb|ZP_00096533.1| hypothetical protein [Novosphingobium aromatic... 84 1e-15
sp|Q59536|PTRB_MORLA PROTEASE II (OLIGOPEPTIDASE B) gi|1084058|p... 83 3e-15
ref|NP_487951.1| protease II [Nostoc sp. PCC 7120] gi|25289820|p... 80 3e-14
>ref|NP_564567.1| expressed protein; protein id: At1g50380.1, supported by cDNA:
gi_15081691, supported by cDNA: gi_19310464 [Arabidopsis
thaliana] gi|25289812|pir||A96540 hypothetical protein
F14I3.4 [imported] - Arabidopsis thaliana
gi|5734786|gb|AAD50051.1|AC007980_16 Similar to
oligopeptidases [Arabidopsis thaliana]
gi|19310465|gb|AAL84967.1| At1g50380/F14I3_27
[Arabidopsis thaliana]
Length = 710
Score = 121 bits (304), Expect = 7e-27
Identities = 56/67 (83%), Positives = 64/67 (94%)
Frame = -1
Query: 633 TAGLNDPRVMYSEPAKFVAKLRDMKTDDNILLFKCELGAGHFSKSGRFEKLQEDAFTYIF 454
TAGLNDPRVMYSEP K+VAKLR+MKTD+N+LLFKCELGAGHFSKSGRFEKLQEDAFT+ F
Sbjct: 639 TAGLNDPRVMYSEPGKWVAKLREMKTDNNVLLFKCELGAGHFSKSGRFEKLQEDAFTFAF 698
Query: 453 ILKALNM 433
++K L+M
Sbjct: 699 MMKVLDM 705
>ref|NP_799677.1| putative protease [Vibrio parahaemolyticus RIMD 2210633]
gi|28808305|dbj|BAC61510.1| putative protease [Vibrio
parahaemolyticus]
Length = 721
Score = 85.5 bits (210), Expect = 5e-16
Identities = 41/66 (62%), Positives = 50/66 (75%)
Frame = -1
Query: 633 TAGLNDPRVMYSEPAKFVAKLRDMKTDDNILLFKCELGAGHFSKSGRFEKLQEDAFTYIF 454
T GL+D +V Y EP K+VAKLR+MKTDDNILLFK ++ AGH SGRF++L+EDA Y F
Sbjct: 654 TTGLHDSQVQYFEPMKWVAKLREMKTDDNILLFKTDMEAGHGGASGRFKRLKEDALEYAF 713
Query: 453 ILKALN 436
L LN
Sbjct: 714 FLDLLN 719
>gb|ZP_00096533.1| hypothetical protein [Novosphingobium aromaticivorans]
Length = 691
Score = 84.3 bits (207), Expect = 1e-15
Identities = 41/69 (59%), Positives = 52/69 (74%)
Frame = -1
Query: 633 TAGLNDPRVMYSEPAKFVAKLRDMKTDDNILLFKCELGAGHFSKSGRFEKLQEDAFTYIF 454
TAGLNDPRV Y EPAK+VA+LR++KTD+N L+ K +GAGH KSGRFE L+E A + F
Sbjct: 623 TAGLNDPRVTYWEPAKWVARLRELKTDENELILKTNMGAGHGGKSGRFESLKETAEEFAF 682
Query: 453 ILKALNMTS 427
IL L + +
Sbjct: 683 ILWQLGVAA 691
>sp|Q59536|PTRB_MORLA PROTEASE II (OLIGOPEPTIDASE B) gi|1084058|pir||JC4185 proteinase II
(EC 3.4.21.-) - Moraxella lacunata
gi|551625|dbj|BAA07460.1| protease II [Moraxella
lacunata]
Length = 690
Score = 83.2 bits (204), Expect = 3e-15
Identities = 40/67 (59%), Positives = 50/67 (73%)
Frame = -1
Query: 633 TAGLNDPRVMYSEPAKFVAKLRDMKTDDNILLFKCELGAGHFSKSGRFEKLQEDAFTYIF 454
T G+NDPRV Y EPAK+VA+LR +KTD+N L+ K +GAGHF KSGRF L+E A +Y F
Sbjct: 614 TTGINDPRVGYFEPAKWVARLRAVKTDNNTLVMKTNMGAGHFGKSGRFNHLKEAAESYAF 673
Query: 453 ILKALNM 433
IL L +
Sbjct: 674 ILDKLGV 680
>ref|NP_487951.1| protease II [Nostoc sp. PCC 7120] gi|25289820|pir||AH2294
proteinase II [imported] - Nostoc sp. (strain PCC 7120)
gi|17133045|dbj|BAB75610.1| protease II [Nostoc sp. PCC
7120]
Length = 688
Score = 79.7 bits (195), Expect = 3e-14
Identities = 39/67 (58%), Positives = 48/67 (71%)
Frame = -1
Query: 633 TAGLNDPRVMYSEPAKFVAKLRDMKTDDNILLFKCELGAGHFSKSGRFEKLQEDAFTYIF 454
TAGLND RV Y EPAK+ AKLR++KTD ++LL K + AGH SGR+E L+E AF Y F
Sbjct: 622 TAGLNDSRVKYWEPAKWTAKLRELKTDHHVLLLKTNMDAGHSGASGRYESLRELAFEYAF 681
Query: 453 ILKALNM 433
IL L +
Sbjct: 682 ILDRLGL 688
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 485,958,823
Number of Sequences: 1393205
Number of extensions: 9368957
Number of successful extensions: 14819
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 14533
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 14808
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26154777244
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)