Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010868A_C01 KMC010868A_c01
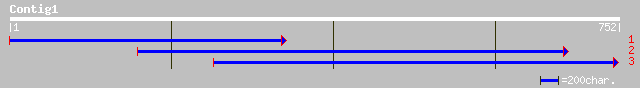
(751 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_200898.1| S-receptor kinase homolog 2 precursor; protein ... 89 7e-17
gb|AAC16898.1| receptor-like protein kinase [Arabidopsis thaliana] 89 7e-17
pir||S27754 S-receptor kinase (EC 2.7.1.-) homolog 2 precursor -... 89 7e-17
ref|NP_181720.1| putative receptor-like protein kinase; protein ... 63 5e-09
gb|AAA32858.1| receptor-like protein kinase 60 4e-08
>ref|NP_200898.1| S-receptor kinase homolog 2 precursor; protein id: At5g60900.1
[Arabidopsis thaliana]
Length = 748
Score = 89.0 bits (219), Expect = 7e-17
Identities = 39/81 (48%), Positives = 58/81 (71%)
Frame = -2
Query: 738 EEKGILSDWAYDCYRSKRLDILLENDNDAAMDTKSFEKFVMIAIWCIQEDPFLRPTMKEV 559
E+ IL +WAYDC+R RL+ L E+D++A D ++ E++V IAIWCIQE+ +RP M+ V
Sbjct: 652 EDNVILINWAYDCFRQGRLEDLTEDDSEAMNDMETVERYVKIAIWCIQEEHGMRPNMRNV 711
Query: 558 LHMLEGIVEVTIPPSPYLYGS 496
MLEG+++V PP+P Y +
Sbjct: 712 TQMLEGVIQVFDPPNPSPYST 732
>gb|AAC16898.1| receptor-like protein kinase [Arabidopsis thaliana]
Length = 100
Score = 89.0 bits (219), Expect = 7e-17
Identities = 39/81 (48%), Positives = 58/81 (71%)
Frame = -2
Query: 738 EEKGILSDWAYDCYRSKRLDILLENDNDAAMDTKSFEKFVMIAIWCIQEDPFLRPTMKEV 559
E+ IL +WAYDC+R RL+ L E+D++A D ++ E++V IAIWCIQE+ +RP M+ V
Sbjct: 4 EDNVILINWAYDCFRQGRLEDLTEDDSEAMNDMETVERYVKIAIWCIQEEHGMRPNMRNV 63
Query: 558 LHMLEGIVEVTIPPSPYLYGS 496
MLEG+++V PP+P Y +
Sbjct: 64 TQMLEGVIQVFDPPNPSPYST 84
>pir||S27754 S-receptor kinase (EC 2.7.1.-) homolog 2 precursor - Arabidopsis
thaliana gi|166846|gb|AAA32857.1| receptor-like protein
kinase
Length = 832
Score = 89.0 bits (219), Expect = 7e-17
Identities = 39/81 (48%), Positives = 58/81 (71%)
Frame = -2
Query: 738 EEKGILSDWAYDCYRSKRLDILLENDNDAAMDTKSFEKFVMIAIWCIQEDPFLRPTMKEV 559
E+ IL +WAYDC+R RL+ L E+D++A D ++ E++V IAIWCIQE+ +RP M+ V
Sbjct: 736 EDNVILINWAYDCFRQGRLEDLTEDDSEAMNDMETVERYVKIAIWCIQEEHGMRPNMRNV 795
Query: 558 LHMLEGIVEVTIPPSPYLYGS 496
MLEG+++V PP+P Y +
Sbjct: 796 TQMLEGVIQVFDPPNPSPYST 816
>ref|NP_181720.1| putative receptor-like protein kinase; protein id: At2g41890.1
[Arabidopsis thaliana] gi|25408762|pir||D84847 probable
receptor-like protein kinase [imported] - Arabidopsis
thaliana gi|1871193|gb|AAB63553.1| putative
receptor-like protein kinase [Arabidopsis thaliana]
gi|20196890|gb|AAM14823.1| putative receptor-like
protein kinase [Arabidopsis thaliana]
Length = 764
Score = 62.8 bits (151), Expect = 5e-09
Identities = 26/77 (33%), Positives = 46/77 (58%)
Frame = -2
Query: 738 EEKGILSDWAYDCYRSKRLDILLENDNDAAMDTKSFEKFVMIAIWCIQEDPFLRPTMKEV 559
E +G++S+W Y + R + +++ + D + E+ + I+ WC+Q D LRP+M EV
Sbjct: 668 EPEGVVSEWVYREWIGGRKETVVDKGLEGCFDVEELERVLRISFWCVQTDERLRPSMGEV 727
Query: 558 LHMLEGIVEVTIPPSPY 508
+ +LEG + V PP P+
Sbjct: 728 VKVLEGTLSVDPPPPPF 744
>gb|AAA32858.1| receptor-like protein kinase
Length = 797
Score = 59.7 bits (143), Expect = 4e-08
Identities = 28/79 (35%), Positives = 45/79 (56%)
Frame = -2
Query: 741 DEEKGILSDWAYDCYRSKRLDILLENDNDAAMDTKSFEKFVMIAIWCIQEDPFLRPTMKE 562
+ EK WA +D ++++ + +T+ + +AIWCIQ++ +RP M
Sbjct: 674 EPEKWFFPPWAAREIIQGNVDSVVDSRLNGEYNTEEVTRMATVAIWCIQDNEEIRPAMGT 733
Query: 561 VLHMLEGIVEVTIPPSPYL 505
V+ MLEG+VEVT+PP P L
Sbjct: 734 VVKMLEGVVEVTVPPPPKL 752
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 632,887,876
Number of Sequences: 1393205
Number of extensions: 13625603
Number of successful extensions: 24605
Number of sequences better than 10.0: 400
Number of HSP's better than 10.0 without gapping: 23742
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24592
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 36314099463
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)