Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010782A_C01 KMC010782A_c01
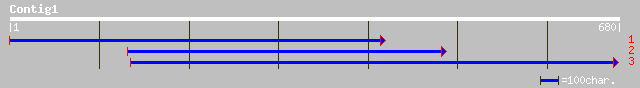
(680 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB92928.1| putative vacuolar proton-ATPase subunit 1 [Oryza... 47 2e-04
gb|AAL78104.1|AC093568_14 Putative proton pump [Oryza sativa] 45 0.001
dbj|BAC41321.1| unnamed protein product [Lotus japonicus] 38 0.11
ref|NP_700610.1| Initiation factor 2 subunit family, putative [P... 38 0.15
emb|CAD27718.1| putative vacuolar ATPase subunit 100 kDa subunit... 38 0.15
>dbj|BAB92928.1| putative vacuolar proton-ATPase subunit 1 [Oryza sativa (japonica
cultivar-group)]
Length = 792
Score = 47.4 bits (111), Expect = 2e-04
Identities = 19/24 (79%), Positives = 21/24 (87%)
Frame = -3
Query: 678 NKFYHGDGYKFKPLSFASLIEDED 607
NKFYHGDGYKF+P SFA L +DED
Sbjct: 769 NKFYHGDGYKFRPFSFALLADDED 792
>gb|AAL78104.1|AC093568_14 Putative proton pump [Oryza sativa]
Length = 783
Score = 44.7 bits (104), Expect = 0.001
Identities = 18/24 (75%), Positives = 21/24 (87%)
Frame = -3
Query: 678 NKFYHGDGYKFKPLSFASLIEDED 607
NKFY GDGYKF P +FAS+IE+ED
Sbjct: 760 NKFYEGDGYKFVPFAFASIIEEED 783
>dbj|BAC41321.1| unnamed protein product [Lotus japonicus]
Length = 702
Score = 38.1 bits (87), Expect = 0.11
Identities = 17/24 (70%), Positives = 20/24 (82%)
Frame = -3
Query: 678 NKFYHGDGYKFKPLSFASLIEDED 607
NKFY GDGYKF P SF SL+++ED
Sbjct: 677 NKFYEGDGYKFFPFSF-SLLDEED 699
>ref|NP_700610.1| Initiation factor 2 subunit family, putative [Plasmodium falciparum
3D7] gi|23495001|gb|AAN35334.1|AE014831_10 Initiation
factor 2 subunit family, putative [Plasmodium falciparum
3D7]
Length = 1074
Score = 37.7 bits (86), Expect = 0.15
Identities = 25/90 (27%), Positives = 41/90 (44%), Gaps = 11/90 (12%)
Frame = +1
Query: 28 NSILLSLFSMQ----------YIYIYMFHARKLPHLSD*TPLKKSKKNMGTMTHGKKITS 177
N IL+ FSM Y+Y++ +H PH D +K KN+ TH K +
Sbjct: 564 NKILIDSFSMNNINNNLDIYDYMYMHHYHHHSNPHKCD----QKCNKNVEGGTHFNKKLN 619
Query: 178 KLSHAYFPLGRSPMCV*SVNTNQSKP-IRY 264
H++ + + + ++NT KP +RY
Sbjct: 620 NFIHSFSDIKKKNILFNNINTKYKKPTVRY 649
>emb|CAD27718.1| putative vacuolar ATPase subunit 100 kDa subunit [Mesembryanthemum
crystallinum]
Length = 816
Score = 37.7 bits (86), Expect = 0.15
Identities = 15/24 (62%), Positives = 20/24 (82%)
Frame = -3
Query: 678 NKFYHGDGYKFKPLSFASLIEDED 607
NKFY GDGYKF P SF+++ E+E+
Sbjct: 793 NKFYLGDGYKFYPFSFSTIGEEEE 816
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 591,938,840
Number of Sequences: 1393205
Number of extensions: 13483163
Number of successful extensions: 29596
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 28352
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29528
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30270070164
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)