Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010713A_C01 KMC010713A_c01
(531 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
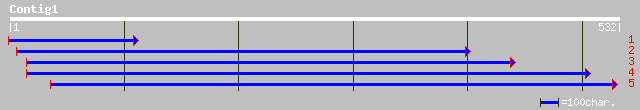
emb|CAD59409.1| SMC1 protein [Oryza sativa] 130 7e-30
ref|NP_191027.1| structural maintenance of chromosomes (SMC) - l... 125 4e-28
gb|AAH46691.1| Similar to SMC-like 1 (yeast) [Xenopus laevis] 76 2e-13
sp|O93308|SMC1_XENLA Structural maintenance of chromosome 1 prot... 76 2e-13
sp|Q14683|SM1A_HUMAN Structural maintenance of chromosome 1-like... 74 8e-13
>emb|CAD59409.1| SMC1 protein [Oryza sativa]
Length = 1264
Score = 130 bits (328), Expect = 7e-30
Identities = 66/81 (81%), Positives = 72/81 (88%)
Frame = -1
Query: 531 DEVDAALDNLNVAKVAGFIRSKSCEGRASQDTDGGSGFQSIVISLKDSFYDKAEALVGVY 352
DEVDAALDNLNVAKVAGFIRSKSC+ R + +GG GFQSIVISLKDSFYDKAEALVGVY
Sbjct: 1185 DEVDAALDNLNVAKVAGFIRSKSCQ-RVDEQDNGGCGFQSIVISLKDSFYDKAEALVGVY 1243
Query: 351 RDSDGGCSKTLTFDLTNYRES 289
RDS+ CS+TLTFDLT YRE+
Sbjct: 1244 RDSERSCSRTLTFDLTKYREA 1264
>ref|NP_191027.1| structural maintenance of chromosomes (SMC) - like protein; protein
id: At3g54670.1 [Arabidopsis thaliana]
gi|11358861|pir||T47626 structural maintenance of
chromosomes (SMC)-like protein - Arabidopsis thaliana
gi|7258371|emb|CAB77587.1| structural maintenance of
chromosomes (SMC)-like protein [Arabidopsis thaliana]
Length = 1265
Score = 125 bits (313), Expect = 4e-28
Identities = 63/82 (76%), Positives = 72/82 (86%), Gaps = 1/82 (1%)
Frame = -1
Query: 531 DEVDAALDNLNVAKVAGFIRSKSCEG-RASQDTDGGSGFQSIVISLKDSFYDKAEALVGV 355
DEVDAALDNLNVAKVA FIRSKSC+ R +QD + G+GFQSIVISLKDSFYDKAEALVGV
Sbjct: 1184 DEVDAALDNLNVAKVAKFIRSKSCQAARDNQDAEDGNGFQSIVISLKDSFYDKAEALVGV 1243
Query: 354 YRDSDGGCSKTLTFDLTNYRES 289
YRD++ CS T++FDL NY+ES
Sbjct: 1244 YRDTERSCSSTMSFDLRNYQES 1265
>gb|AAH46691.1| Similar to SMC-like 1 (yeast) [Xenopus laevis]
Length = 1232
Score = 76.3 bits (186), Expect = 2e-13
Identities = 43/83 (51%), Positives = 55/83 (65%), Gaps = 2/83 (2%)
Frame = -1
Query: 531 DEVDAALDNLNVAKVAGFIRSKSCEGRASQDTDGGSGFQSIVISLKDSFYDKAEALVGVY 352
DE+DAALDN N+ KVA +I+ +S S FQ+IVISLK+ FY KAE+L+GVY
Sbjct: 1156 DEIDAALDNTNIGKVANYIKEQSM-----------SNFQAIVISLKEEFYTKAESLIGVY 1204
Query: 351 RDSDGGC--SKTLTFDLTNYRES 289
+ G C SK LTFDLT Y ++
Sbjct: 1205 PE-QGDCVISKVLTFDLTKYPDA 1226
>sp|O93308|SMC1_XENLA Structural maintenance of chromosome 1 protein
gi|3328231|gb|AAC26807.1| 14S cohesin SMC1 subunit; SMC
protein [Xenopus laevis]
Length = 1232
Score = 76.3 bits (186), Expect = 2e-13
Identities = 43/83 (51%), Positives = 55/83 (65%), Gaps = 2/83 (2%)
Frame = -1
Query: 531 DEVDAALDNLNVAKVAGFIRSKSCEGRASQDTDGGSGFQSIVISLKDSFYDKAEALVGVY 352
DE+DAALDN N+ KVA +I+ +S S FQ+IVISLK+ FY KAE+L+GVY
Sbjct: 1156 DEIDAALDNTNIGKVANYIKEQSM-----------SNFQAIVISLKEEFYTKAESLIGVY 1204
Query: 351 RDSDGGC--SKTLTFDLTNYRES 289
+ G C SK LTFDLT Y ++
Sbjct: 1205 PE-QGDCVISKVLTFDLTKYPDA 1226
>sp|Q14683|SM1A_HUMAN Structural maintenance of chromosome 1-like 1 protein (SMC1alpha
protein) (SB1.8/DXS423E protein) (Sb1.8)
gi|20521836|dbj|BAA11495.2| KIAA0178 [Homo sapiens]
Length = 1233
Score = 74.3 bits (181), Expect = 8e-13
Identities = 42/83 (50%), Positives = 54/83 (64%), Gaps = 2/83 (2%)
Frame = -1
Query: 531 DEVDAALDNLNVAKVAGFIRSKSCEGRASQDTDGGSGFQSIVISLKDSFYDKAEALVGVY 352
DE+DAALDN N+ KVA +I+ +S FQ+IVISLK+ FY KAE+L+GVY
Sbjct: 1156 DEIDAALDNTNIGKVANYIKEQST-----------CNFQAIVISLKEEFYTKAESLIGVY 1204
Query: 351 RDSDGGC--SKTLTFDLTNYRES 289
+ G C SK LTFDLT Y ++
Sbjct: 1205 PE-QGDCVISKVLTFDLTKYPDA 1226
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 411,558,751
Number of Sequences: 1393205
Number of extensions: 8326553
Number of successful extensions: 16476
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 16112
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 16405
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17596710992
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)