Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
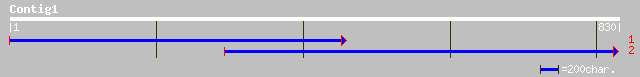
Query= KMC010657A_C01 KMC010657A_c01
(830 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_178361.1| hypothetical protein; protein id: At2g02570.1 [... 91 2e-17
gb|AAO22752.1| unknown protein [Arabidopsis thaliana] gi|2839390... 91 2e-17
gb|EAA17439.1| hypothetical protein [Plasmodium yoelii yoelii] 40 0.042
gb|EAA31184.1| predicted protein [Neurospora crassa] 37 0.27
gb|AAF45352.1| CG17454 gene product [Drosophila melanogaster] 37 0.46
>ref|NP_178361.1| hypothetical protein; protein id: At2g02570.1 [Arabidopsis
thaliana] gi|7487808|pir||T00606 hypothetical protein
At2g02570 [imported] - Arabidopsis thaliana
gi|3184282|gb|AAC18929.1| hypothetical protein
[Arabidopsis thaliana]
Length = 316
Score = 91.3 bits (225), Expect = 2e-17
Identities = 40/54 (74%), Positives = 48/54 (88%)
Frame = -3
Query: 660 QIGFFSGRKRESIFKSPDDPQGKVGVTGSGKGLTEFQKREKHFHLKDGSVENDE 499
++GFF+GRK+ESIFKSP+DP GKVGVTGSGKGLT+FQKREKH HLK G+ E +
Sbjct: 262 KVGFFTGRKKESIFKSPEDPFGKVGVTGSGKGLTDFQKREKHLHLKSGNAEGTD 315
>gb|AAO22752.1| unknown protein [Arabidopsis thaliana] gi|28393903|gb|AAO42359.1|
unknown protein [Arabidopsis thaliana]
Length = 300
Score = 91.3 bits (225), Expect = 2e-17
Identities = 40/54 (74%), Positives = 48/54 (88%)
Frame = -3
Query: 660 QIGFFSGRKRESIFKSPDDPQGKVGVTGSGKGLTEFQKREKHFHLKDGSVENDE 499
++GFF+GRK+ESIFKSP+DP GKVGVTGSGKGLT+FQKREKH HLK G+ E +
Sbjct: 246 KVGFFTGRKKESIFKSPEDPFGKVGVTGSGKGLTDFQKREKHLHLKSGNAEGTD 299
>gb|EAA17439.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 5309
Score = 40.0 bits (92), Expect = 0.042
Identities = 30/88 (34%), Positives = 43/88 (48%), Gaps = 10/88 (11%)
Frame = +3
Query: 75 NYDFLSSKETN*FIHNAK*VILRGNFCEVSFENLS*NLATSNLTQE----------NLPL 224
N+D LS +N IHN K L GNF + + ENL+ +SNL +E N +
Sbjct: 4775 NHDSLSESNSNDSIHNNKEENLDGNFSKNNQENLT----SSNLDEEKKEVIDENIFNSIV 4830
Query: 225 QETLNLSHKSQQNLQHHNKLR*TNKIGL 308
+ L K + NL H NK + N+I +
Sbjct: 4831 YKIAKLFKKKKSNLNHDNKKKKKNEINM 4858
>gb|EAA31184.1| predicted protein [Neurospora crassa]
Length = 370
Score = 37.4 bits (85), Expect = 0.27
Identities = 14/36 (38%), Positives = 25/36 (68%)
Frame = -3
Query: 636 KRESIFKSPDDPQGKVGVTGSGKGLTEFQKREKHFH 529
K+ES+F++P+ G+VG TGSG+ + + R +H +
Sbjct: 328 KKESMFRTPEGVNGRVGFTGSGQAMRKDPTRSRHIY 363
>gb|AAF45352.1| CG17454 gene product [Drosophila melanogaster]
Length = 240
Score = 36.6 bits (83), Expect = 0.46
Identities = 19/40 (47%), Positives = 26/40 (64%), Gaps = 3/40 (7%)
Frame = -3
Query: 645 SGRKRESIFKSPDDPQGKVGV---TGSGKGLTEFQKREKH 535
+G K SIF SPD+ G+VGV +GKG+T+F EK+
Sbjct: 197 NGMKARSIFASPDNVSGRVGVGTCGTAGKGMTDFTVGEKY 236
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 657,011,020
Number of Sequences: 1393205
Number of extensions: 13542418
Number of successful extensions: 25458
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 24721
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25455
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 42922608498
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)