Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010480A_C01 KMC010480A_c01
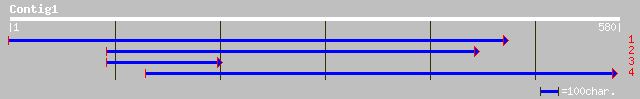
(577 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T47604 oligopeptide transporter-like protein - Arabidopsis ... 87 3e-22
ref|NP_173670.1| oligopeptide transporter, putative; protein id:... 78 6e-19
gb|AAL57662.1| At1g72120/F28P5_2 [Arabidopsis thaliana] gi|24111... 69 2e-17
dbj|BAB64117.1| P0039A07.25 [Oryza sativa (japonica cultivar-gro... 74 3e-17
gb|AAO18439.1| putative peptide transport protein [Oryza sativa ... 77 7e-17
>pir||T47604 oligopeptide transporter-like protein - Arabidopsis thaliana
gi|7258348|emb|CAB77565.1| oligopeptide transporter-like
protein [Arabidopsis thaliana]
Length = 555
Score = 87.4 bits (215), Expect(3) = 3e-22
Identities = 38/62 (61%), Positives = 50/62 (80%)
Frame = -3
Query: 575 WLLPQYTITGVSDAFPIVGLQELFYDPMPESLRSLGAAAYISLVGVGSFASNVVIAVVEA 396
WLLPQY + G+ D F IVG+QELFYD MPE++RS+GAA +IS+VGVGSF S +I+ V+
Sbjct: 436 WLLPQYILVGIGDVFTIVGMQELFYDQMPETMRSIGAAIFISVVGVGSFVSTGIISTVQT 495
Query: 395 VS 390
+S
Sbjct: 496 IS 497
Score = 36.2 bits (82), Expect(3) = 3e-22
Identities = 15/34 (44%), Positives = 25/34 (73%)
Frame = -2
Query: 348 RPPLDGFYWVMAVMSAVHLGAYLWLAKAFVYQKV 247
R LD +YW++A ++AV L YL++A F+Y+K+
Sbjct: 511 RAHLDYYYWIIASLNAVSLCFYLFIANHFLYKKL 544
Score = 23.1 bits (48), Expect(3) = 3e-22
Identities = 9/12 (75%), Positives = 10/12 (83%)
Frame = -1
Query: 379 GEKWLGNNLNPA 344
GE+WL NNLN A
Sbjct: 501 GEEWLVNNLNRA 512
>ref|NP_173670.1| oligopeptide transporter, putative; protein id: At1g22540.1
[Arabidopsis thaliana] gi|25402970|pir||F86358 Similar
to peptide transporter [imported] - Arabidopsis thaliana
gi|6587835|gb|AAF18524.1|AC006551_10 Similar to peptide
transporter [Arabidopsis thaliana]
Length = 570
Score = 77.8 bits (190), Expect(2) = 6e-19
Identities = 31/64 (48%), Positives = 49/64 (76%)
Frame = -3
Query: 575 WLLPQYTITGVSDAFPIVGLQELFYDPMPESLRSLGAAAYISLVGVGSFASNVVIAVVEA 396
WL+PQY + G++D F +VGLQE FYD +P LRS+G A Y+S+ G+G+F S+ +I+++E
Sbjct: 458 WLVPQYVLFGITDVFAMVGLQEFFYDQVPNELRSVGLALYLSIFGIGNFLSSFMISIIEK 517
Query: 395 VSSR 384
+S+
Sbjct: 518 ATSQ 521
Score = 37.7 bits (86), Expect(2) = 6e-19
Identities = 14/32 (43%), Positives = 26/32 (80%)
Frame = -2
Query: 339 LDGFYWVMAVMSAVHLGAYLWLAKAFVYQKVD 244
LD FYW++A +S + L +YL++AK++V +++D
Sbjct: 537 LDYFYWLLACLSFIGLASYLYVAKSYVSKRLD 568
>gb|AAL57662.1| At1g72120/F28P5_2 [Arabidopsis thaliana] gi|24111301|gb|AAN46774.1|
At1g72120/F28P5_2 [Arabidopsis thaliana]
Length = 557
Score = 69.3 bits (168), Expect(2) = 2e-17
Identities = 27/59 (45%), Positives = 45/59 (75%)
Frame = -3
Query: 575 WLLPQYTITGVSDAFPIVGLQELFYDPMPESLRSLGAAAYISLVGVGSFASNVVIAVVE 399
WL+PQY + G++D + +VG+QE FY +P LRS+G A Y+S +GVGS S+++I++++
Sbjct: 445 WLIPQYLLLGLADVYTLVGMQEFFYSQVPTELRSIGLALYLSALGVGSLLSSLLISLID 503
Score = 40.8 bits (94), Expect(2) = 2e-17
Identities = 15/35 (42%), Positives = 28/35 (79%)
Frame = -2
Query: 348 RPPLDGFYWVMAVMSAVHLGAYLWLAKAFVYQKVD 244
R LD FYW++A++SAV +L+++K+++Y++VD
Sbjct: 521 RAHLDYFYWLLAIVSAVGFFTFLFISKSYIYRRVD 555
>dbj|BAB64117.1| P0039A07.25 [Oryza sativa (japonica cultivar-group)]
gi|19571117|dbj|BAB86541.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
gi|20146489|dbj|BAB89268.1| P0491F11.1 [Oryza sativa
(japonica cultivar-group)]
Length = 617
Score = 74.3 bits (181), Expect(2) = 3e-17
Identities = 29/64 (45%), Positives = 48/64 (74%)
Frame = -3
Query: 575 WLLPQYTITGVSDAFPIVGLQELFYDPMPESLRSLGAAAYISLVGVGSFASNVVIAVVEA 396
W++PQY + G+SD F ++GLQE FYD +P++LRSLG A ++S+ GVG F S+ +I+ ++
Sbjct: 500 WMVPQYVLFGLSDVFAMIGLQEFFYDQVPDALRSLGLAFFLSIFGVGHFFSSFIISAIDG 559
Query: 395 VSSR 384
+ +
Sbjct: 560 ATKK 563
Score = 35.4 bits (80), Expect(2) = 3e-17
Identities = 14/33 (42%), Positives = 25/33 (75%)
Frame = -2
Query: 348 RPPLDGFYWVMAVMSAVHLGAYLWLAKAFVYQK 250
R LD FYW++A + AV L A++++++ +VY+K
Sbjct: 575 RAHLDYFYWLLAGLCAVELVAFVFVSRVYVYKK 607
>gb|AAO18439.1| putative peptide transport protein [Oryza sativa (japonica
cultivar-group)]
Length = 585
Score = 76.6 bits (187), Expect(2) = 7e-17
Identities = 32/63 (50%), Positives = 47/63 (73%)
Frame = -3
Query: 575 WLLPQYTITGVSDAFPIVGLQELFYDPMPESLRSLGAAAYISLVGVGSFASNVVIAVVEA 396
WL+PQ+ + G+ D F +VGLQE FYD +P+S+RSLG Y+S++G GSF S+ +I V+
Sbjct: 450 WLVPQFLLMGIGDGFALVGLQEYFYDQVPDSMRSLGIGLYLSVIGAGSFLSSQLITAVDR 509
Query: 395 VSS 387
V+S
Sbjct: 510 VTS 512
Score = 32.0 bits (71), Expect(2) = 7e-17
Identities = 13/37 (35%), Positives = 23/37 (62%)
Frame = -2
Query: 339 LDGFYWVMAVMSAVHLGAYLWLAKAFVYQKVDGGGEI 229
LD FYW++A + +L Y+ +A + Y+ V GG++
Sbjct: 530 LDLFYWLLACIGVANLVFYVVIATRYSYKTVMAGGKV 566
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 550,483,316
Number of Sequences: 1393205
Number of extensions: 13402351
Number of successful extensions: 48249
Number of sequences better than 10.0: 191
Number of HSP's better than 10.0 without gapping: 45262
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 48127
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21530810025
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)