Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010467A_C01 KMC010467A_c01
(606 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
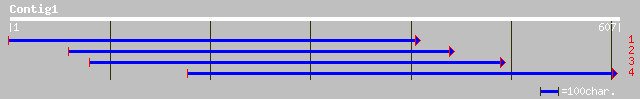
ref|XP_139214.1| hypothetical protein XP_139214 [Mus musculus] 35 0.76
sp|P02851|BAR2_CHITE BALBIANI RING PROTEIN 2 (GIANT SECRETORY PR... 35 0.99
ref|NP_378192.1| 630aa long hypothetical protein [Sulfolobus tok... 34 1.3
ref|NP_056891.1| Pr180 [Mason-Pfizer monkey virus] gi|2801483|gb... 34 1.3
sp|P07572|POL_MPMV POL polyprotein [Contains: Reverse transcript... 34 1.3
>ref|XP_139214.1| hypothetical protein XP_139214 [Mus musculus]
Length = 399
Score = 35.0 bits (79), Expect = 0.76
Identities = 31/102 (30%), Positives = 38/102 (36%), Gaps = 18/102 (17%)
Frame = +3
Query: 165 FATQCQCKMSYRITCTSVQTPAINSKSIHYQHRSLLHH-----------------LKPSN 293
+ CQC + C S TP + + S+ H SLLH L S
Sbjct: 168 YEMNCQCSYTIAPYCISAVTPLLLTASVQLHHCSLLHQCSYTIAPYCISAATPLLLTASV 227
Query: 294 SSFHCVLKSLHTIS-TYLPQ*IVVAVPIL*TVQCQHFYISPL 416
HC L LH S T P I A P+L T Q + S L
Sbjct: 228 QLHHCSL--LHQCSYTIAPYCISAATPLLLTASVQLHHCSLL 267
>sp|P02851|BAR2_CHITE BALBIANI RING PROTEIN 2 (GIANT SECRETORY PROTEIN I-B) (GSP-IB)
gi|72279|pir||BQIC2 Balbiani ring-2 chain - midge
(Chironomus tentans) (fragment) gi|7097|emb|CAA23476.1|
Balbiani gene 2 product [Chironomus tentans]
Length = 210
Score = 34.7 bits (78), Expect = 0.99
Identities = 31/93 (33%), Positives = 42/93 (44%), Gaps = 2/93 (2%)
Frame = +3
Query: 27 SKNNKKQKKFHKQKPTL-ALPSTKSK*NRIESRIKQNTWKKC*NKDEFATQCQCKMSYRI 203
SK++K K KP+ + PS SK + S +K+ KC K+ +C
Sbjct: 1 SKHSKPSKHSKHSKPSKHSKPSKHSKPEKCGSAMKRTEAAKCARKNGRFNSKRC------ 54
Query: 204 TCTSVQTPAINSK-SIHYQHRSLLHHLKPSNSS 299
TCTSV P+ SK S +H H KPS S
Sbjct: 55 TCTSVGKPSKPSKHSKPSKHSKPSKHSKPSKHS 87
Score = 34.3 bits (77), Expect = 1.3
Identities = 31/95 (32%), Positives = 42/95 (43%), Gaps = 2/95 (2%)
Frame = +3
Query: 21 RRSKNNKKQKKFHKQKPTL-ALPSTKSK*NRIESRIKQNTWKKC*NKDEFATQCQCKMSY 197
+ SK +K K KP+ + PS SK + S +K+ KC K+ +C
Sbjct: 73 KHSKPSKHSKPSKHSKPSKHSKPSKHSKPEKCGSAMKRTEAAKCARKNGRFNSKRC---- 128
Query: 198 RITCTSVQTPAINSK-SIHYQHRSLLHHLKPSNSS 299
TCTSV P+ SK S +H H KPS S
Sbjct: 129 --TCTSVGKPSKPSKHSKPSKHSKPSKHSKPSKHS 161
>ref|NP_378192.1| 630aa long hypothetical protein [Sulfolobus tokodaii]
gi|15623313|dbj|BAB67301.1| 630aa long hypothetical
protein [Sulfolobus tokodaii]
Length = 630
Score = 34.3 bits (77), Expect = 1.3
Identities = 24/80 (30%), Positives = 39/80 (48%), Gaps = 6/80 (7%)
Frame = +2
Query: 245 NSLSAQKLASSPKTVQLFIPLCAQIP------SYYIHISTSMNSSSSTHPLNCTVSTLLH 406
+SL + L SS TV +P+ AQ+ YY + + S+SST + T +
Sbjct: 387 SSLISSSLTSSSGTVSFTLPISAQVKVNVSAFGYYPNSTVVTVSASSTVKIYLTPIQITV 446
Query: 407 ISTTSSLNSAPFLEFQINPT 466
I TT ++N + QI+P+
Sbjct: 447 IPTTFTINGSKITPEQISPS 466
>ref|NP_056891.1| Pr180 [Mason-Pfizer monkey virus] gi|2801483|gb|AAC82576.1| Pr180
[Mason-Pfizer monkey virus]
Length = 1771
Score = 34.3 bits (77), Expect = 1.3
Identities = 24/73 (32%), Positives = 38/73 (51%)
Frame = +2
Query: 209 YKCTDTSNQFQINSLSAQKLASSPKTVQLFIPLCAQIPSYYIHISTSMNSSSSTHPLNCT 388
Y TDT+ +FQ N SAQ + +Q I + + P+ ++I T + + PL T
Sbjct: 1379 YTLTDTTIKFQTNLNSAQLV-----ELQALIAVLSAFPNQPLNIYTDSAYLAHSIPLLET 1433
Query: 389 VSTLLHISTTSSL 427
V+ + HIS T+ L
Sbjct: 1434 VAQIKHISETAKL 1446
>sp|P07572|POL_MPMV POL polyprotein [Contains: Reverse transcriptase ; Endonuclease]
gi|74616|pir||GNLJMP pol polyprotein (clone 6A) -
Mason-Pfizer monkey virus gi|334704|gb|AAA47711.1| pol
polyprotein
Length = 867
Score = 34.3 bits (77), Expect = 1.3
Identities = 24/73 (32%), Positives = 38/73 (51%)
Frame = +2
Query: 209 YKCTDTSNQFQINSLSAQKLASSPKTVQLFIPLCAQIPSYYIHISTSMNSSSSTHPLNCT 388
Y TDT+ +FQ N SAQ + +Q I + + P+ ++I T + + PL T
Sbjct: 475 YTLTDTTIKFQTNLNSAQLV-----ELQALIAVLSAFPNQPLNIYTDSAYLAHSIPLLET 529
Query: 389 VSTLLHISTTSSL 427
V+ + HIS T+ L
Sbjct: 530 VAQIKHISETAKL 542
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 544,544,269
Number of Sequences: 1393205
Number of extensions: 12083658
Number of successful extensions: 31401
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 29899
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31316
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23997478008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)